3B1P
 
 | | Structure of Burkholderia thailandensis nucleoside kinase (BthNK) in complex with ADP-inosine | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSINE, Ribokinase, ... | | Authors: | Yasutake, Y, Ota, H, Hino, E, Sakasegawa, S, Tamura, T. | | Deposit date: | 2011-07-05 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Burkholderia thailandensis nucleoside kinase: implications for the catalytic mechanism and nucleoside selectivity
Acta Crystallogr.,Sect.D, 67, 2011
|
|
7M5L
 
 | | PCNA bound to peptide mimetic with linker | | Descriptor: | PROPANE, Peptide mimetic (ACE)RQCSMTCFYHSK(NH2) with linker, Proliferating cell nuclear antigen | | Authors: | Vandborg, B.A, Bruning, J.B. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A cell permeable bimane-constrained PCNA-interacting peptide.
Rsc Chem Biol, 2, 2021
|
|
3SKC
 
 | |
4MFJ
 
 | | The crystal structure of acyltransferase | | Descriptor: | Putative uncharacterized protein tcp24 | | Authors: | Lyu, S.Y, Liu, Y.C, Chang, C.Y, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
7M5N
 
 | | PCNA bound to peptide mimetic with linker | | Descriptor: | 1,3-dimethylbenzene, Peptide mimetic (ACE)RQCSMTCFYHSK(NH2) with linker, Proliferating cell nuclear antigen | | Authors: | Vandborg, B.A, Bruning, J.B. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | A cell permeable bimane-constrained PCNA-interacting peptide.
Rsc Chem Biol, 2, 2021
|
|
7MDZ
 
 | | 80S rabbit ribosome stalled with benzamide-CHX | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S21, ... | | Authors: | Koga, Y, Hoang, E.M, Park, Y, Keszei, A.F.A, Murray, J, Shao, S, Liau, B.B. | | Deposit date: | 2021-04-06 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery of C13-Aminobenzoyl Cycloheximide Derivatives that Potently Inhibit Translation Elongation.
J.Am.Chem.Soc., 143, 2021
|
|
4M2R
 
 | | Human Carbonic Anhydrase II in complex with Brinzolamide | | Descriptor: | (+)-4-ETHYLAMINO-3,4-DIHYDRO-2-(METHOXY)PROPYL-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Pinard, M.P, Boone, C.D, Rife, B.D, Supuran, C.T, Mckenna, R. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | Structural study of interaction between brinzolamide and dorzolamide inhibition of human carbonic anhydrases.
Bioorg.Med.Chem., 21, 2013
|
|
3SAP
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[(5-butyl-2-pyrimidinyl)sulfanyl]acetyl}benzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-{[(5-butylpyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, BICINE, ... | | Authors: | Grazulis, S, Manakova, E, Tamulaitiene, G. | | Deposit date: | 2011-06-03 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design of [(2-pyrimidinylthio)acetyl]benzenesulfonamides as inhibitors of human carbonic anhydrases.
Eur.J.Med.Chem., 51, 2012
|
|
3SAX
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2-chloro-5-{[(5-ethyl-2-pyrimidinyl)sulfanyl]acetyl}benzenesulfonamide | | Descriptor: | 2-chloro-5-{[(5-ethylpyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Grazulis, S, Manakova, E, Tamulaitiene, G. | | Deposit date: | 2011-06-03 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Design of [(2-pyrimidinylthio)acetyl]benzenesulfonamides as inhibitors of human carbonic anhydrases.
Eur.J.Med.Chem., 51, 2012
|
|
4MFL
 
 | | The crystal structure of acyltransferase in complex with decanoyl-CoA and Tei pseudoaglycone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, Putative uncharacterized protein tcp24, ... | | Authors: | Lyu, S.Y, Liu, Y.C, Chang, C.Y, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
3SBI
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-[(2-pyrimidinylsulfanyl)acetyl]benzenesulfonamide | | Descriptor: | 4-[(pyrimidin-2-ylsulfanyl)acetyl]benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Grazulis, S, Manakova, E, Tamulaitiene, G. | | Deposit date: | 2011-06-05 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design of [(2-pyrimidinylthio)acetyl]benzenesulfonamides as inhibitors of human carbonic anhydrases.
Eur.J.Med.Chem., 51, 2012
|
|
3B7Z
 
 | | Crystal Structure of Yeast Sec14 Homolog Sfh1 in Complex with Phosphatidylcholine or Phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Uncharacterized protein YKL091C | | Authors: | Ortlund, E.A, Schaaf, G, Redinbo, M.R, Bankaitis, V. | | Deposit date: | 2007-10-31 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily
Mol.Cell, 29, 2008
|
|
2DX3
 
 | |
4MBU
 
 | |
7XNY
 
 | |
7XNX
 
 | |
4MFQ
 
 | | The crystal structure of acyltransferase in complex with CoA and 10C-Teicoplanin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, COENZYME A, ... | | Authors: | Lyu, S.Y, Liu, Y.C, Chang, C.Y, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
3S8X
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[(4-methyl-6-oxo-1,6-dihydro-2-pyrimidinyl)sulfanyl]acetyl}benzenesulfonamide | | Descriptor: | 4-{[(4-methyl-6-oxo-1,6-dihydropyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Grazulis, S, Manakova, E, Tamulaitiene, G. | | Deposit date: | 2011-05-31 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design of [(2-pyrimidinylthio)acetyl]benzenesulfonamides as inhibitors of human carbonic anhydrases.
Eur.J.Med.Chem., 51, 2012
|
|
5M62
 
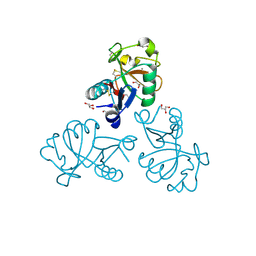 | | Structure of the Mus musclus Langerin carbohydrate recognition domain in complex with glucose | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, GLYCEROL, ... | | Authors: | Loll, B, Aretz, J, Rademacher, C, Wahl, M.C. | | Deposit date: | 2016-10-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bacterial Polysaccharide Specificity of the Pattern Recognition Receptor Langerin Is Highly Species-dependent.
J. Biol. Chem., 292, 2017
|
|
3BF3
 
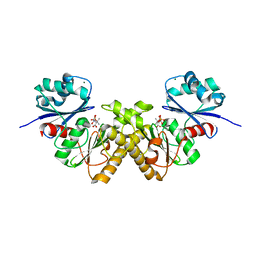 | | Type III pantothenate kinase from Thermotoga maritima complexed with product phosphopantothenate | | Descriptor: | MAGNESIUM ION, N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanine, Type III pantothenate kinase | | Authors: | Yang, K, Huerta, C, Strauss, E, Zhang, H. | | Deposit date: | 2007-11-20 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for substrate binding and the catalytic mechanism of type III pantothenate kinase.
Biochemistry, 47, 2008
|
|
5MQY
 
 | | CATHEPSIN L IN COMPLEX WITH 4-[1,3-benzodioxol-5-ylmethyl(2-phenoxyethyl)amino]-5-fluoropyrimidine-2-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, 4-[1,3-benzodioxol-5-ylmethyl(2-phenoxyethyl)amino]-5-fluoropyrimidine-2-carbonitrile, Cathepsin L1 | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-12-21 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Prospective Evaluation of Free Energy Calculations for the Prioritization of Cathepsin L Inhibitors.
J. Med. Chem., 60, 2017
|
|
7MH9
 
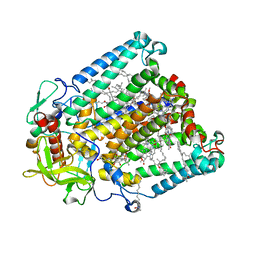 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-nitrotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH4
 
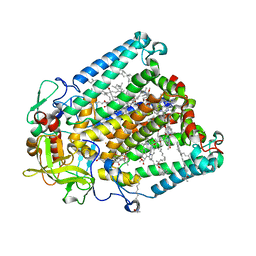 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-bromotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH5
 
 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-iodotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH8
 
 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-methyltyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
