7M3U
 
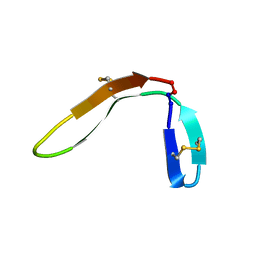 | |
7MMY
 
 | |
3KNE
 
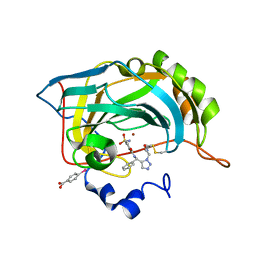 | | Carbonic Anhydrase II H64C mutant in complex with an in situ formed triazole | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, Carbonic anhydrase 2, N-[(S)-(1-{2-oxo-2-[(3-sulfanylpropyl)amino]ethyl}-1H-1,2,3-triazol-5-yl)(phenyl)methyl]-4-sulfamoylbenzamide, ... | | Authors: | Schulze-Wischeler, J, Heine, A, Klebe, G. | | Deposit date: | 2009-11-12 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Stereo- and Regioselective Azide/Alkyne Cycloadditions in Carbonic Anhydrase II via Tethering, Monitored by Crystallography and Mass Spectrometry.
Chemistry, 17, 2011
|
|
6UJA
 
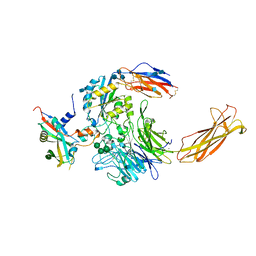 | | Integrin alpha-v beta-8 in complex with pro-TGF-beta1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Campbell, M.G, Cormier, A, Cheng, Y, Nishimura, S.L. | | Deposit date: | 2019-10-02 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Reveals Integrin-Mediated TGF-beta Activation without Release from Latent TGF-beta.
Cell, 180, 2020
|
|
3KSZ
 
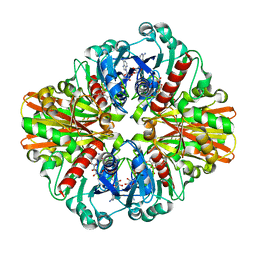 | | Crystal Structure of C151S+H178N mutant of Glyceraldehyde-3-phosphate-dehydrogenase 1 (GAPDH 1) from Staphylococcus aureus MRSA252 complexed with NAD and G3P | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-11-24 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
8J9D
 
 | | Crystal structure of M61 peptidase (bestatin-bound) from Xanthomonas campestris | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Yadav, P, Kumar, A, Kulkarni, B.S, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a newly identified M61 family aminopeptidase with broad substrate specificity that is solely responsible for recycling acidic amino acids.
Febs J., 291, 2024
|
|
3KRB
 
 | |
2Q9U
 
 | | Crystal structure of the flavodiiron protein from Giardia intestinalis | | Descriptor: | A-type flavoprotein, FLAVIN MONONUCLEOTIDE, MU-OXO-DIIRON, ... | | Authors: | Di Matteo, A, Scandurra, F.M, Testa, F, Forte, E, Sarti, P, Brunori, M, Giuffre, A. | | Deposit date: | 2007-06-14 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The O2-scavenging flavodiiron protein in the human parasite Giardia intestinalis
J.Biol.Chem., 283, 2008
|
|
7X3V
 
 | | Cryo-EM structure of IOC3-N2 nucleosome | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Lifei, L, Kangjing, C, Chen, Z. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8KD5
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD4
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD6
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD2
 
 | | Rpd3S in complex with 187bp nucleosome | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD7
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 167bp DNA | | Descriptor: | 167bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
6UEL
 
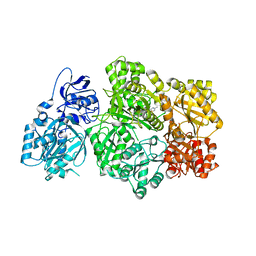 | | CPS1 bound to allosteric inhibitor H3B-193 | | Descriptor: | Carbamoyl-phosphate synthase [ammonia], mitochondrial, N~1~-[(4-fluorophenyl)methyl]-N~1~-methyl-N~4~-(4-methyl-1,3-thiazol-2-yl)piperidine-1,4-dicarboxamide, ... | | Authors: | Larsen, N.A, Nguyen, T.V. | | Deposit date: | 2019-09-21 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small Molecule Inhibition of CPS1 Activity through an Allosteric Pocket.
Cell Chem Biol, 27, 2020
|
|
5YU4
 
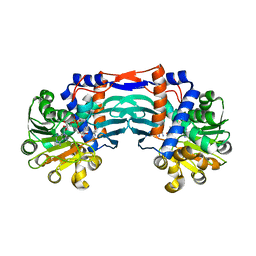 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5YU0
 
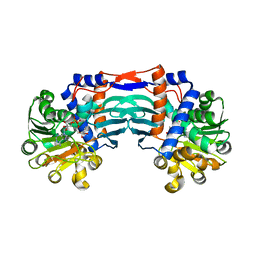 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | Descriptor: | Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
4FMO
 
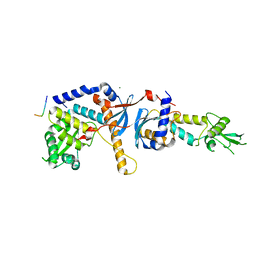 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer bound to a fragment of exo1 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, DNA repair peptide, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-06-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of the MutL alpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
5YU1
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | Descriptor: | (2S)-piperidine-2-carboxylic acid, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
4FMN
 
 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer bound to a fragment of NTG2 | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-06-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of the MutL alpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
5YU3
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | Descriptor: | Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROLINE, ... | | Authors: | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
4E4W
 
 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-03-13 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the MutLalpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3NWC
 
 | |
4MFK
 
 | | The crystal structure of acyltransferase in complex with decanoyl-CoA | | Descriptor: | Putative uncharacterized protein tcp24, SULFATE ION, decanoyl-CoA | | Authors: | Lyu, S.Y, Liu, Y.C, Chang, C.Y, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
4MB2
 
 | | Crystal structure of TON1374 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Phosphopantothenate synthetase | | Authors: | Kim, M.-K, An, Y.J, Cha, S.-S. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The crystal structure of a novel phosphopantothenate synthetase from the hyperthermophilic archaea, Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 439, 2013
|
|
