6T2X
 
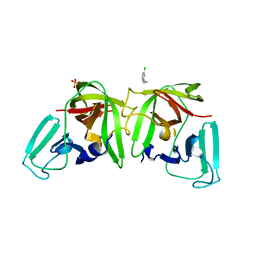 | |
6T3G
 
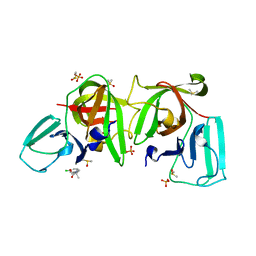 | |
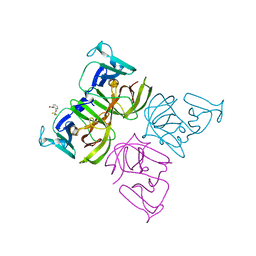
6T4S
 
 | |
6T5D
 
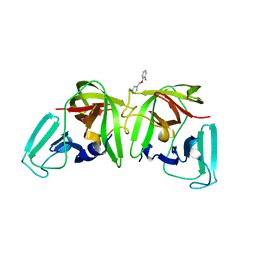 | |
6TAL
 
 | | 3C-like protease from Southampton virus complexed with FMOPL000227a. | | Descriptor: | 5-ethyl-1,3,4-thiadiazol-2-amine, DIMETHYL SULFOXIDE, Genome polyprotein, ... | | Authors: | Guo, J, Cooper, J.B. | | Deposit date: | 2019-10-29 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | In crystallo-screening for discovery of human norovirus 3C-like protease inhibitors.
J Struct Biol X, 4, 2020
|
|
7PBL
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s1 [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBO
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s4 [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBP
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s5 [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBM
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s2 [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBN
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s3 [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
3EN3
 
 | | Crystal Structure of the GluR4 Ligand-Binding domain in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 4,Glutamate receptor | | Authors: | Gill, A, Madden, D.R. | | Deposit date: | 2008-09-25 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Correlating AMPA receptor activation and cleft closure across subunits: crystal structures of the GluR4 ligand-binding domain in complex with full and partial agonists
Biochemistry, 47, 2008
|
|
3FWG
 
 | | Ferric camphor bound Cytochrome P450cam, Arg365Leu, Glu366Gln, monoclinic crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Camphor 5-monooxygenase, ... | | Authors: | Schlichting, I, Von Koenig, K, Aldag, C, Hilvert, D. | | Deposit date: | 2009-01-18 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Probing the role of the proximal heme ligand in cytochrome P450cam by recombinant incorporation of selenocysteine.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8TCF
 
 | | Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Campbell, M.G, Fernandez, A, Roy, A, Kraft, J, Baker, D. | | Deposit date: | 2023-06-30 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
8RKH
 
 | | Crystal structure of the ZP-N2 and ZP-N3 domains of mouse ZP2 (mZP2-N2N3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2, ... | | Authors: | Fahrenkamp, D, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8RKE
 
 | | Crystal structure of the complete N-terminal region of human ZP2 (hZP2-N1N2N3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fahrenkamp, D, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
3EPE
 
 | |
2LFR
 
 | | Solution structure of the chimeric Af1503 HAMP- EnvZ DHp homodimer | | Descriptor: | HAMP domain-containing protein, Osmolarity sensor protein EnzV chimera | | Authors: | Coles, M, Ferris, H.U, Hulko, M, Martin, J, Lupas, A.N. | | Deposit date: | 2011-07-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism of regulation of receptor histidine kinases.
Structure, 20, 2012
|
|
2LFS
 
 | | Solution structure of the chimeric Af1503 HAMP- EnvZ DHp homodimer; A219F variant | | Descriptor: | HAMP domain-containing protein, Osmolarity sensor protein EnzV chimera | | Authors: | Coles, M, Ferris, H.U, Hulko, M, Martin, J, Lupas, A.N. | | Deposit date: | 2011-07-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism of regulation of receptor histidine kinases.
Structure, 20, 2012
|
|
6I7F
 
 | | Dehaloperoxidase B from Amphitrite ornata - complex with 2,4-dichlorophenol | | Descriptor: | 2,4-dichlorophenol, Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Moreno-Chicano, T, Ebrahim, A, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Sugimoto, H, Tono, K, Owada, S, Duyvesteyn, H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-throughput structures of protein-ligand complexes at room temperature using serial femtosecond crystallography.
Iucrj, 6, 2019
|
|
6O9G
 
 | | Open state GluA2 in complex with STZ and blocked by AgTx-636, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2019-03-13 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
6I6G
 
 | | Dehaloperoxidase B from Amphitrite ornata - complex with 5-bromoindole | | Descriptor: | 5-bromanyl-1~{H}-indole, Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Moreno-Chicano, T, Ebrahim, A, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Sugimoto, H, Tono, K, Owada, S, Duyvesteyn, H. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-20 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-throughput structures of protein-ligand complexes at room temperature using serial femtosecond crystallography.
Iucrj, 6, 2019
|
|
2L9G
 
 | |
6DLZ
 
 | | Open state GluA2 in complex with STZ after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
6DM1
 
 | | Open state GluA2 in complex with STZ and blocked by NASPM, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
6DM0
 
 | | Open state GluA2 in complex with STZ and blocked by IEM-1460, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
