8PZ8
 
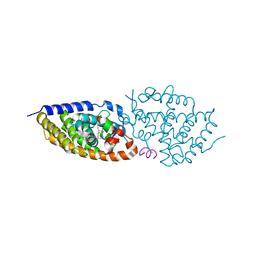 | | crystal structure of VDR in complex with D-Bishomo-1a,25-dihydroxyvitamin D3 Analog 54 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(4~{a}~{R},5~{S},9~{a}~{S})-4~{a}-methyl-5-[(2~{R})-6-methyl-6-oxidanyl-heptan-2-yl]-3,4,5,6,7,8,9,9~{a}-octahydro-2~{H}-benzo[7]annulen-1-ylidene]ethylidene]cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2023-07-27 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Design, synthesis, and biological activity of D-bishomo-1 alpha ,25-dihydroxyvitamin D 3 analogs and their crystal structures with the vitamin D nuclear receptor.
Eur.J.Med.Chem., 271, 2024
|
|
1M8W
 
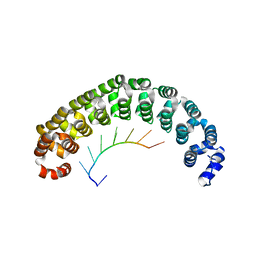 | | CRYSTAL STRUCTURE OF THE PUMILIO-HOMOLOGY DOMAIN FROM HUMAN PUMILIO1 IN COMPLEX WITH NRE1-19 RNA | | Descriptor: | 5'-R(P*UP*GP*UP*AP*UP*AP*U)-3', 5'-R(P*UP*GP*UP*CP*CP*AP*G)-3', 5'-R(P*UP*UP*GP*UP*AP*UP*AP*U)-3', ... | | Authors: | Wang, X, McLachlan, J, Zamore, P.D, Hall, T.M.T. | | Deposit date: | 2002-07-26 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MODULAR RECOGNITION OF RNA BY A HUMAN PUMILIO-HOMOLOGY DOMAIN
CELL(CAMBRIDGE,MASS.), 110, 2002
|
|
8PZ6
 
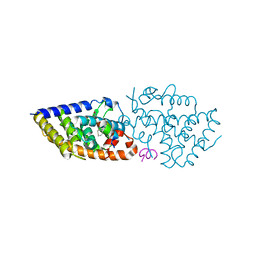 | | crystal structure of VDR in complex with D-Bishomo-1a,25-dihydroxyvitamin D3 analog 56 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(4~{a}~{R},5~{S},9~{a}~{S})-4~{a}-methyl-5-[(2~{R})-6-methyl-6-oxidanyl-heptan-2-yl]-3,4,5,6,7,8,9,9~{a}-octahydro-2~{H}-benzo[7]annulen-1-ylidene]ethylidene]-2-(3-oxidanylpropylidene)cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2023-07-27 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological activity of D-bishomo-1 alpha ,25-dihydroxyvitamin D 3 analogs and their crystal structures with the vitamin D nuclear receptor.
Eur.J.Med.Chem., 271, 2024
|
|
1MFQ
 
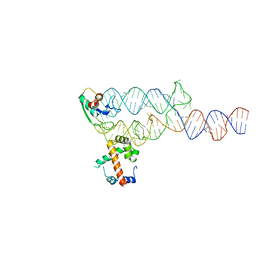 | | Crystal Structure Analysis of a Ternary S-Domain Complex of Human Signal Recognition Particle | | Descriptor: | 7S RNA of human SRP, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kuglstatter, A, Oubridge, C, Nagai, K. | | Deposit date: | 2002-08-13 | | Release date: | 2002-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Induced structural changes of 7SL RNA during the assembly of human
signal recognition particle
Nat.Struct.Biol., 9, 2002
|
|
2ZBE
 
 | |
1M5K
 
 | |
8HHQ
 
 | |
1NI3
 
 | |
8FPE
 
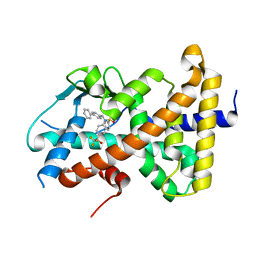 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-BP | | Descriptor: | N-[([1,1'-biphenyl]-4-yl)methyl]-N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Li, Y, Chen, T. | | Deposit date: | 2023-01-04 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2XPJ
 
 | |
6M37
 
 | |
2QLZ
 
 | |
6M36
 
 | |
1NJI
 
 | | Structure of chloramphenicol bound to the 50S ribosomal subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10e, 50S ribosomal protein L13P, ... | | Authors: | Hansen, J.L, Moore, P.B, Steitz, T.A. | | Deposit date: | 2002-12-31 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of Five Antibiotics Bound at the Peptidyl Transferase Center of
the Large Ribosomal Subunit
J.Mol.Biol., 330, 2003
|
|
8H5B
 
 | | The cryo-EM structure of nuclear transport receptor Kap114p complex with yeast TATA-box binding protein | | Descriptor: | Importin subunit beta-5, TATA-box-binding protein | | Authors: | Hsia, K.C, Liao, C.C, Wang, C.H, Wu, Y.M. | | Deposit date: | 2022-10-12 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural convergence endows nuclear transport receptor Kap114p with a transcriptional repressor function toward TATA-binding protein.
Nat Commun, 14, 2023
|
|
6SOD
 
 | | Fragment N14056a in complex with MAP kinase p38-alpha | | Descriptor: | 1-[[(3~{S})-1,4-dioxaspiro[4.5]decan-3-yl]methyl]piperidine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Nichols, C.E, De Nicola, G.F. | | Deposit date: | 2019-08-29 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
3R00
 
 | |
1NXZ
 
 | | X-Ray Crystal Structure of Protein yggj_haein of Haemophilus influenzae. Northeast Structural Genomics Consortium Target IR73. | | Descriptor: | Hypothetical protein HI0303 | | Authors: | Forouhar, F, Shen, J, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional assignment based on structural analysis: crystal structure of the yggJ protein (HI0303) of Haemophilus influenzae reveals an RNA methyltransferase with a deep trefoil knot.
Proteins, 53, 2003
|
|
5YP5
 
 | | Crystal structure of RORgamma complexed with SRC2 and compound 5d | | Descriptor: | 2-[4-(ethylsulfonyl)phenyl]-N-{5-[2-(2-methylpropyl)benzoyl]-4-phenyl-1,3-thiazol-2-yl}acetamide, Nuclear receptor ROR-gamma, SRC2-2 peptide | | Authors: | Gao, M, Cai, W, Chunwa, C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | From ROR gamma t Agonist to Two Types of ROR gamma t Inverse Agonists
ACS Med Chem Lett, 9, 2018
|
|
6X02
 
 | |
6X03
 
 | |
1NYR
 
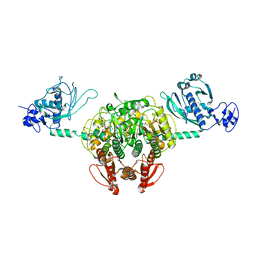 | | Structure of Staphylococcus aureus threonyl-tRNA synthetase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, THREONINE, ZINC ION, ... | | Authors: | Torres-Larios, A, Sankaranarayanan, R, Rees, B, Dock-Bregeon, A.C, Moras, D. | | Deposit date: | 2003-02-13 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational movements and cooperativity upon amino acid, ATP and tRNA binding in threonyl-tRNA synthetase
J.Mol.Biol., 331, 2003
|
|
6XFV
 
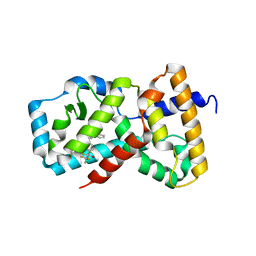 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C IN COMPLEX WITH A NOVEL INVERSE AGONIST | | Descriptor: | 1-(4-{(3S,4S)-4-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-3-methyl-3-phenylpyrrolidine-1-carbonyl}piperidin-1-yl)ethan-1-one, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2020-06-16 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of (3S,4S)-3-methyl-3-(4-fluorophenyl)-4-(4-(1,1,1,3,3,3-hexafluoro-2-hydroxyprop-2-yl)phenyl)pyrrolidines as novel ROR gamma t inverse agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6SOV
 
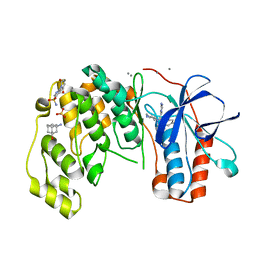 | | Fragments KCL_615 and KCL_802 in complex with MAP kinase p38-alpha | | Descriptor: | (5~{S},7~{R})-3-azanyladamantan-1-ol, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, 6-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]pyridine-3-sulfonamide, ... | | Authors: | Nichols, C.E, De Nicola, G.F. | | Deposit date: | 2019-08-30 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
1O03
 
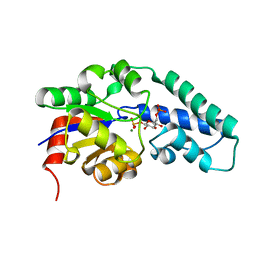 | | Structure of Pentavalent Phosphorous Intermediate of an Enzyme Catalyzed Phosphoryl transfer Reaction observed on cocrystallization with Glucose 6-phosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-02-20 | | Release date: | 2003-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The pentacovalent phosphorus intermediate of a phosphoryl transfer reaction.
Science, 299, 2003
|
|
