5A8I
 
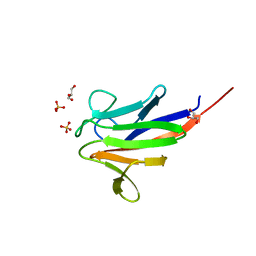 | | Crystal structure of the FHA domain of ArnA from Sulfolobus acidocaldarius | | Descriptor: | ARNA, GLYCEROL, SULFATE ION | | Authors: | Hoffmann, L, Anders, K, Reimann, J, Linne, U, Essen, L.-O, Albers, S.-V. | | Deposit date: | 2015-07-16 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phosphorylation-Dependent Interaction between an Archaeal Von Willebrand and Fha Domain Recruits Arna-Arnb Complex to DNA for Repression of Motility
To be Published
|
|
3FOH
 
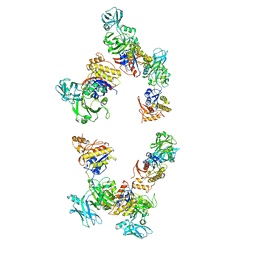 | | Fitting of gp18M crystal structure into 3D cryo-EM reconstruction of bacteriophage T4 extended tail | | Descriptor: | Tail sheath protein Gp18 | | Authors: | Aksyuk, A.A, Leiman, P.G, Kurochkina, L.P, Shneider, M.M, Kostyuchenko, V.A, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2008-12-30 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria.
Embo J., 28, 2009
|
|
4GM3
 
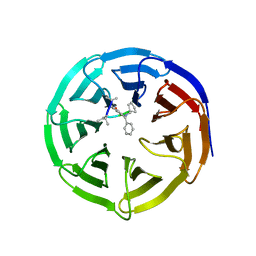 | | Crystal structure of human WD repeat domain 5 with compound MM-101 | | Descriptor: | MM-101, WD repeat-containing protein 5 | | Authors: | Karatas, H, Townsend, E.C, Chen, Y, Bernard, D, Cao, F, Liu, L, Lei, M, Dou, Y, Wang, S. | | Deposit date: | 2012-08-15 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.393 Å) | | Cite: | High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction.
J.Am.Chem.Soc., 135, 2013
|
|
7JVB
 
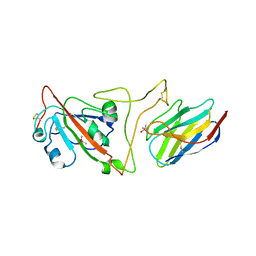 | | Crystal structure of the SARS-CoV-2 spike receptor-binding domain (RBD) with nanobody Nb20 | | Descriptor: | CACODYLATE ION, Nanobody Nb20, Spike protein S1 | | Authors: | Xiang, Y, Xiao, Z, Liu, H, Sang, Z, Schneidman-Duhovny, D, Zhang, C, Shi, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.287 Å) | | Cite: | Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2.
Science, 370, 2020
|
|
3FJF
 
 | |
7QSP
 
 | | Permutated C-terminal lobe of the ribose binding protein from Thermotoga maritima | | Descriptor: | 1,2-ETHANEDIOL, Ribose ABC transporter, periplasmic ribose-binding protein | | Authors: | Shanmugaratnam, S, Michel, F, Hocker, B. | | Deposit date: | 2022-01-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of permuted halves of a modern ribose-binding protein.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
2BXG
 
 | | Human serum albumin complexed with ibuprofen | | Descriptor: | IBUPROFEN, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
2X86
 
 | | AGME bound to ADP-B-mannose | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADP-L-GLYCERO-D-MANNO-HEPTOSE-6-EPIMERASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kowatz, T, Morrison, J.P, Tanner, M.E, Naismith, J.H. | | Deposit date: | 2010-03-06 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the Y140F Mutant of Adp-L-Glycero-D-Manno-Heptose 6-Epimerase Bound to Adp-Beta-D-Mannose Suggests a One Base Mechanism.
Protein Sci., 19, 2010
|
|
7JVO
 
 | |
7QNS
 
 | | Peptide VYEKKP in complex with human cathepsin V C25S mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Cathepsin L2, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-12-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Peptide VYEKKP in complex with human cathepsin V C25S mutant
To Be Published
|
|
2WK2
 
 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitotrio-thiazoline dithioamide. | | Descriptor: | 2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose-(1-4)-2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, CHITINASE A, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-04 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chitinase Inhibition by Chitobiose and Chitotriose Thiazolines.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
1PHA
 
 | | INHIBITOR-INDUCED CONFORMATIONAL CHANGE IN CYTOCHROME P450-CAM | | Descriptor: | 1-(N-IMIDAZOLYL)-2-HYDROXY-2-(2,3-DICHLOROPHENYL)OCTANE, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Poulos, T.L. | | Deposit date: | 1992-07-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Inhibitor-induced conformational change in cytochrome P-450CAM.
Biochemistry, 32, 1993
|
|
4N1B
 
 | | STRUCTURE OF KEAP1 KELCH DOMAIN WITH(1S,2R)-2-[(1S)-1-[(1-oxo-2,3-dihydro-1H-isoindol-2-Yl)methyl]-1,2,3,4-tetrahydroisoquinoline-2-Carbonyl]cyclohexane-1-carboxylic acid | | Descriptor: | (1S,2R)-2-{[(1S)-1-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Smith, M.A, Duclos, S, Beaumont, E, Kwong, J, Brooks, M, Barker, J, Jnoff, E, Brookfield, F, Courade, J.P, Barker, O, Fryatt, T, Albrecht, C, Bromidge, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-02-19 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
7K19
 
 | | CryoEM structure of DNA-PK catalytic subunit complexed with DNA (Complex I) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
2ZCH
 
 | | Crystal structure of human prostate specific antigen complexed with an activating antibody | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Menez, R, Stura, E, Jolivet-Reynaud, C. | | Deposit date: | 2007-11-08 | | Release date: | 2008-01-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of a ternary complex between human prostate-specific antigen, its substrate acyl intermediate and an activating antibody
J.Mol.Biol., 376, 2008
|
|
4FUA
 
 | | L-FUCULOSE-1-PHOSPHATE ALDOLASE COMPLEX WITH PGH | | Descriptor: | BETA-MERCAPTOETHANOL, L-FUCULOSE-1-PHOSPHATE ALDOLASE, PHOSPHOGLYCOLOHYDROXAMIC ACID, ... | | Authors: | Dreyer, M.K, Schulz, G.E. | | Deposit date: | 1996-02-14 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Catalytic mechanism of the metal-dependent fuculose aldolase from Escherichia coli as derived from the structure.
J.Mol.Biol., 259, 1996
|
|
2ED2
 
 | | Solution Structure of RSGI RUH-069, a GTF2I domain in human cDNA | | Descriptor: | General transcription factor II-I | | Authors: | Doi-Katayama, Y, Hirota, H, Izumi, K, Hayashi, F, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-14 | | Release date: | 2007-08-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-069, a GTF2I domain in human cDNA
To be published
|
|
2X56
 
 | | Yersinia Pestis Plasminogen Activator Pla (Native) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, COAGULASE/FIBRINOLYSIN | | Authors: | Eren, E, Murphy, M, Goguen, J, van den Berg, B. | | Deposit date: | 2010-02-05 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Active Site Water Network in the Plasminogen Activator Pla from Yersinia Pestis
Structure, 18, 2010
|
|
8WP9
 
 | | Small-heat shock protein from Methanocaldococcus jannaschii, Hsp16.5 | | Descriptor: | Small heat shock protein HSP16.5 | | Authors: | Lee, J, Ryu, B, Kim, T, Kim, K.K. | | Deposit date: | 2023-10-09 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structure of a 16.5-kDa small heat-shock protein from Methanocaldococcus jannaschii.
Int.J.Biol.Macromol., 258, 2024
|
|
7JY9
 
 | | Structure of a 9 base pair RecA-D loop complex | | Descriptor: | DNA (27-MER), DNA (42-MER), MAGNESIUM ION, ... | | Authors: | Pavletich, N.P. | | Deposit date: | 2020-08-29 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of strand exchange from RecA-DNA synaptic and D-loop structures.
Nature, 586, 2020
|
|
5A2G
 
 | | An esterase from anaerobic Clostridium hathewayi can hydrolyze aliphatic aromatic polyesters | | Descriptor: | CARBOXYLIC ESTER HYDROLASE, PHOSPHATE ION | | Authors: | Hromic, A, Pavkov Keller, T, Steinkellner, G, Gruber, K, Perz, V, Baumschlager, A, Bleymaier, K, Zitzenbacher, S, Zankel, A, Mayrhofer, C, Sinkel, C, Kueper, U, Schlegel, K.A, Ribitsch, D, Guebitz, G.M. | | Deposit date: | 2015-05-19 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | An Esterase from Anaerobic Clostridium Hathewayi Can Hydrolyze Aliphatic-Aromatic Polyesters.
Environ.Sci.Tech., 50, 2016
|
|
2ZT8
 
 | | Crystal structure of human Glycyl-tRNA synthetase (GlyRS) in complex with Gly-AMP analog | | Descriptor: | Glycyl-tRNA synthetase, [(2S,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (aminoacetyl)sulfamate | | Authors: | Guo, R.T, Yang, X.L, Schimmel, P. | | Deposit date: | 2008-09-19 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structures and biochemical analyses suggest unique mechanism and role for human GlyRS in Ap4A homeostasis
To be Published
|
|
2ZTI
 
 | |
4GPH
 
 | |
1PJD
 
 | | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers | | Descriptor: | Pheromone alpha factor receptor | | Authors: | Valentine, K.G, Liu, S.-F, Marassi, F.M, Veglia, G, Nevzorov, A.A, Opella, S.J, Ding, F.-X, Wang, S.-H, Arshava, B, Becker, J.M, Naider, F. | | Deposit date: | 2003-06-02 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers
Biopolymers, 59, 2001
|
|
