6S88
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(2-Methoxy-5-((1,2,4,5-tetrahydro-3H-benzo[d]azepin-3-yl)sulfonyl)phenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
7FYF
 
 | | Crystal Structure of human FABP4 in complex with 4-(4-fluorophenyl)-2-piperidin-1-yl-3-(1H-tetrazol-5-yl)-7,8-dihydro-5H-pyrano[4,3-b]pyridine, i.e. SMILES c1(ccc(cc1)F)c1c(c(N2CCCCC2)nc2c1COCC2)C1=NN=NN1 with IC50=0.42193 microM | | Descriptor: | (3M)-4-(4-fluorophenyl)-2-(piperidin-1-yl)-3-(1H-tetrazol-5-yl)-7,8-dihydro-5H-pyrano[4,3-b]pyridine, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7G1C
 
 | | Crystal Structure of human FABP4 in complex with rac-(1R,2S)-2-[(3-methylphenoxy)methyl]cyclohexane-1-carboxylic acid, i.e. SMILES C1CC[C@H]([C@H](C1)C(=O)O)COc1cc(ccc1)C with IC50=1.28108 microM | | Descriptor: | (1S,2R)-2-[(3-methylphenoxy)methyl]cyclohexane-1-carboxylic acid, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Buettelmann, B, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FW2
 
 | | Crystal Structure of human FABP4 in complex with 5-[(4-chlorophenoxy)methyl]-4-prop-2-enyl-1,2,4-triazole-3-thiol, i.e. SMILES N1(C(=NN=C1COc1ccc(Cl)cc1)S)CC=C with IC50=0.833 microM | | Descriptor: | 5-[(4-chlorophenoxy)methyl]-4-(prop-2-en-1-yl)-4H-1,2,4-triazole-3-thiol, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5ENZ
 
 | | S. aureus MnaA-UDP co-structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Chemical Genetic Analysis and Functional Characterization of Staphylococcal Wall Teichoic Acid 2-Epimerases Reveals Unconventional Antibiotic Drug Targets.
Plos Pathog., 12, 2016
|
|
5OK0
 
 | | Structure of the D10N mutant of beta-phosphoglucomutase from Lactococcus lactis trapped with native reaction intermediate beta-glucose 1,6-bisphosphate to 2.2A resolution. | | Descriptor: | 1,3-PROPANDIOL, 1,6-di-O-phosphono-beta-D-glucopyranose, Beta-phosphoglucomutase, ... | | Authors: | Robertson, A.J, Bisson, C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | van der Waals Contact between Nucleophile and Transferring Phosphorus Is Insufficient To Achieve Enzyme Transition-State Architecture
Acs Catalysis, 2018
|
|
6WJ1
 
 | | Crystal structure of Fab 54-4H03 bound to H1 influenza hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 54-4H03 heavy chain, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-04-11 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Convergent Evolution in Breadth of Two VH6-1-Encoded Influenza Antibody Clonotypes from a Single Donor.
Cell Host Microbe, 28, 2020
|
|
7AZW
 
 | | Crystal structure of the MIZ1-BTB-domain | | Descriptor: | GLYCEROL, Zinc finger and BTB domain-containing protein 17 isoform X1 | | Authors: | Orth, B, Sander, B, Diederichs, K, Lorenz, S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of an atypical interaction site in the BTB domain of the MYC-interacting zinc-finger protein 1.
Structure, 29, 2021
|
|
6HWO
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-1335 | | Descriptor: | 1,2-ETHANEDIOL, 3-[5-[(4aR,8aS)-4-OXIDANYLIDENE-3-PROPAN-2-YL-4a,5,8,8a-TETRAHYDROPHTHALAZIN-1-YL]-2-METHOXY-PHENYL]-N-(PHENYLMETHYL)PROP-2-YNAMIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-10-12 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors (Part 2).
Bioorg.Med.Chem., 27, 2019
|
|
5HHT
 
 | |
7FXE
 
 | | Crystal Structure of human FABP4 in complex with 2-(9-benzyl-6-methyl-1,2,3,4-tetrahydrocarbazol-1-yl)acetic acid, i.e. SMILES C12=C(c3c(N1Cc1ccccc1)ccc(c3)C)CCC[C@H]2CC(=O)O with IC50=3.60776 microM | | Descriptor: | Fatty acid-binding protein, adipocyte, SULFATE ION, ... | | Authors: | Ehler, A, Benz, J, Obst, U, ROCHE, N, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5EKT
 
 | |
6CSS
 
 | |
5NRW
 
 | |
6RUJ
 
 | |
5I1F
 
 | |
9DBY
 
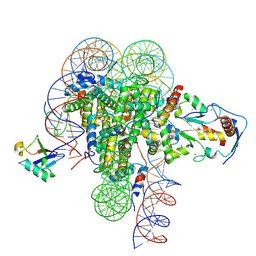 | | ncPRC1RYBP bound to singly modified H2AK119Ub nucleosome | | Descriptor: | DNA (187-MER), E3 ubiquitin-protein ligase RING2, Histone H2A type 1, ... | | Authors: | Godinez-Lopez, V, Valencia-Sanchez, M.I, Armache, J.P, Armache, K.-J. | | Deposit date: | 2024-08-24 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Read-write mechanisms of H2A ubiquitination by Polycomb repressive complex 1.
Nature, 636, 2024
|
|
6H8X
 
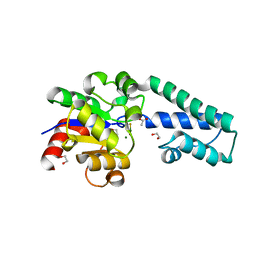 | | Beta-phosphoglucomutase from Lactococcus lactis in an open conformer complexed with magnesium trifluoride to 1.8 A. | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ACETATE ION, ... | | Authors: | Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-03 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
7B7Y
 
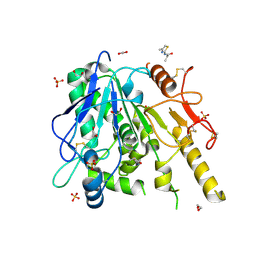 | | Notum-Fragment064 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, N-(4-methyl-1,3-thiazol-2-yl)propanamide, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-11 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Notum Fragment Screen
To Be Published
|
|
7B7X
 
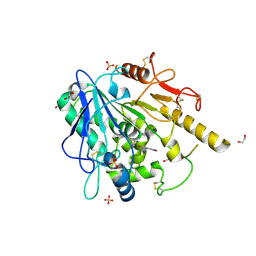 | | Notum-Fragment063 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, N-(2,1,3-benzoxadiazol-4-yl)acetamide, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-11 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Notum Fragment Screen
To Be Published
|
|
7B7W
 
 | | Notum-Fragment049 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, N-ethyl-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-11 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Notum Fragment Screen
To Be Published
|
|
9DDE
 
 | | ncPRC1RYBP bound to H2AK119Ub/H1.4 chromatosome | | Descriptor: | DNA (187-MER), E3 ubiquitin-protein ligase RING2, Histone H1.4, ... | | Authors: | Godinez-Lopez, V, Valencia-Sanchez, M.I, Armache, J.P, Armache, K.-J. | | Deposit date: | 2024-08-28 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Read-write mechanisms of H2A ubiquitination by Polycomb repressive complex 1.
Nature, 636, 2024
|
|
9JS5
 
 | | Crystal structure of the ASFV-derived histone-like protein pA104R | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Viral histone-like protein | | Authors: | Li, Q, Shao, H, Yi, D, Cen, S. | | Deposit date: | 2024-09-30 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thonningianin A disrupts pA104R-DNA binding and inhibits African swine fever virus replication.
Emerg Microbes Infect, 14, 2025
|
|
8WMD
 
 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-2 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-10-03 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
6D15
 
 | | Crystal structure of KPC-2 complexed with compound 1 | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, [(6,7-dimethyl-2-oxo-2H-1-benzopyran-4-yl)methyl]phosphonic acid | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
