3EVM
 
 | | Crystal structure of the Mimivirus NDK +Kpn-N62L-R107G triple mutant complexed with dCDP | | Descriptor: | DEOXYCYTIDINE DIPHOSPHATE, MAGNESIUM ION, Nucleoside diphosphate kinase | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-10-13 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
1IAN
 
 | | HUMAN P38 MAP KINASE INHIBITOR COMPLEX | | Descriptor: | 4-[5-(3-IODO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-1H-IMIDAZOL-4-YL]-PYRIDINE, P38 MAP KINASE | | Authors: | Tong, L. | | Deposit date: | 1997-03-07 | | Release date: | 1998-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A highly specific inhibitor of human p38 MAP kinase binds in the ATP pocket.
Nat.Struct.Biol., 4, 1997
|
|
6U62
 
 | | Raptor-Rag-Ragulator complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rogala, K.B, Sabatini, D.M. | | Deposit date: | 2019-08-29 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis for the docking of mTORC1 on the lysosomal surface.
Science, 366, 2019
|
|
3F0A
 
 | | Structure of a putative n-acetyltransferase (ta0374) in complex with acetyl-coa from thermoplasma acidophilum | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, N-ACETYLTRANSFERASE, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
3F1Q
 
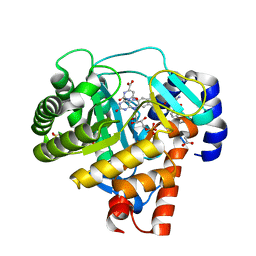 | | Human dihydroorotate dehydrogenase in complex with a leflunomide derivative inhibitor 1 | | Descriptor: | (2Z)-N-biphenyl-4-yl-2-cyano-3-hydroxybut-2-enamide, ACETIC ACID, Dihydroorotate dehydrogenase, ... | | Authors: | Heikkila, T, Davies, M, McConkey, G.A, Fishwick, C.W.G, Parsons, M.R, Johnson, A.P. | | Deposit date: | 2008-10-28 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis, and characterization of inhibitors of human and Plasmodium falciparum dihydroorotate dehydrogenases
J.Med.Chem., 52, 2009
|
|
3X1H
 
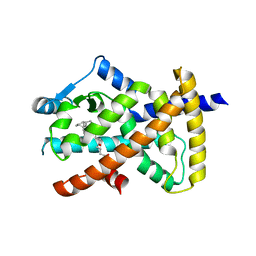 | | hPPARgamma Ligand binding domain in complex with 5-oxo-tricosahexaenoic acid | | Descriptor: | (7E,11Z,14Z,17Z,20Z)-5-oxotricosa-7,11,14,17,20-pentaenoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-11-18 | | Release date: | 2015-04-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of covalent bond formation between PPAR gamma and oxo-fatty acids.
Bioconjug.Chem., 26, 2015
|
|
2WHH
 
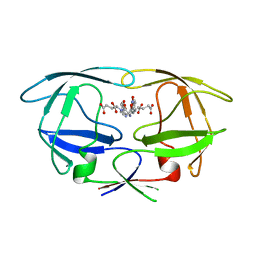 | | HIV-1 protease tethered dimer Q-product complex along with nucleophilic water molecule | | Descriptor: | GLUTAMIC ACID, PARA-NITROPHENYLALANINE, POL PROTEIN | | Authors: | Prashar, V, Bihani, S, Das, A, Ferrer, J.L, Hosur, M.V. | | Deposit date: | 2009-05-05 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Catalytic Water Co-Existing with a Product Peptide in the Active Site of HIV-1 Protease Revealed by X- Ray Structure Analysis.
Plos One, 4, 2009
|
|
3EXV
 
 | |
3X1I
 
 | | hPPARgamma Ligand binding domain in complex with 6-oxo-tetracosahexaenoic acid | | Descriptor: | (8E,12Z,15Z,18Z,21Z)-6-oxotetracosa-8,12,15,18,21-pentaenoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-11-18 | | Release date: | 2015-04-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of covalent bond formation between PPAR gamma and oxo-fatty acids.
Bioconjug.Chem., 26, 2015
|
|
3FCG
 
 | | Crystal Structure Analysis of the Middle Domain of the Caf1A Usher | | Descriptor: | CHLORIDE ION, F1 capsule-anchoring protein | | Authors: | Yu, X, Visweswaran, G.R, Duck, Z, Marupakula, S, MacIntyre, S, Knight, S, Zavialov, A.V. | | Deposit date: | 2008-11-21 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Caf1A usher possesses a Caf1 subunit-like domain that is crucial for Caf1 fibre secretion
Biochem.J., 418, 2009
|
|
5C9Z
 
 | |
6AC8
 
 | | Crystal structure of Mycobacterium smegmatis Mfd at 2.75 A resolution | | Descriptor: | Mycobacterium smegmatis Mfd, SULFATE ION | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6XF9
 
 | | Crystal structure of KSHV ORF68 | | Descriptor: | Packaging protein UL32, ZINC ION | | Authors: | Didychuk, A.L, Gates, S.N, Martin, A, Glaunsinger, B. | | Deposit date: | 2020-06-15 | | Release date: | 2021-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A pentameric protein ring with novel architecture is required for herpesviral packaging.
Elife, 10, 2021
|
|
6XFA
 
 | | Cryo-EM structure of EBV BFLF1 | | Descriptor: | Packaging protein UL32, ZINC ION | | Authors: | Didychuk, A.L, Gates, S.N, Martin, A, Glaunsinger, B. | | Deposit date: | 2020-06-15 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A pentameric protein ring with novel architecture is required for herpesviral packaging.
Elife, 10, 2021
|
|
7R7S
 
 | | p47-bound p97-R155H mutant with ATPgammaS | | Descriptor: | NSFL1 cofactor p47, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7R7T
 
 | | p47-bound p97-R155H mutant with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NSFL1 cofactor p47, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7R7U
 
 | | D1 and D2 domain structure of the p97(R155H)-p47 complex | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7RLH
 
 | | Cryo-EM structure of human p97-D592N mutant bound to ATPgS. | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7RLG
 
 | | Cryo-EM structure of human p97-D592N mutant bound to ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
5U6I
 
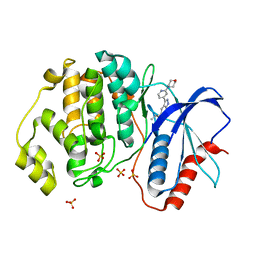 | | Discovery of MLi-2, an Orally Available and Selective LRRK2 Inhibitor that Reduces Brain Kinase Activity | | Descriptor: | 3-[2-(morpholin-4-yl)pyridin-4-yl]-5-[(propan-2-yl)oxy]-1H-indazole, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Scott, J.D, DeMong, D.E, Fell, M.J, Mirescu, C, Basu, K, Greshock, T.J, Morrow, J.A, Xiao, L, Hruza, A, Harris, J, Tiscia, H.E, Chang, R.K, Embrey, M.W, McCauley, J.A, Li, W, Lin, S, Liu, H, Dai, X, Baptista, M, Agnihotri, G, Columbus, J, Mei, H, Poirier, M, Zhou, X, Lin, Y, Yin, Z, Sanders, J.M, Drolet, R.E, Kern, J.T, Kennedy, M.E, Parker, E.M, Stamford, A.W, Nargund, R, Miller, M.W. | | Deposit date: | 2016-12-08 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of a 3-(4-Pyrimidinyl) Indazole (MLi-2), an Orally Available and Selective Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitor that Reduces Brain Kinase Activity.
J. Med. Chem., 60, 2017
|
|
7XAY
 
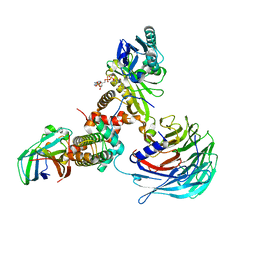 | | Crystal structure of Hat1-Hat2-Asf1-H3-H4 | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yue, Y, Yang, W.S, Xu, R.M. | | Deposit date: | 2022-03-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Topography of histone H3-H4 interaction with the Hat1-Hat2 acetyltransferase complex.
Genes Dev., 36, 2022
|
|
6X4A
 
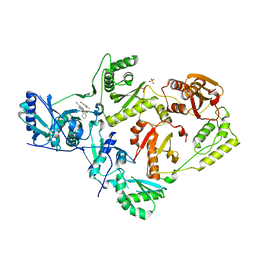 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile (JLJ651), a Non-nucleoside Inhibitor | | Descriptor: | 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.537 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
4G56
 
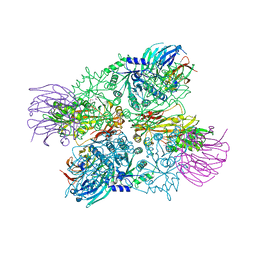 | | Crystal Structure of full length PRMT5/MEP50 complexes from Xenopus laevis | | Descriptor: | Hsl7 protein, MGC81050 protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Ho, M, Wilczek, C, Bonanno, J, Shechter, D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity
Plos One, 8, 2013
|
|
7RLC
 
 | | Cryo-EM structure of human p97-A232E mutant bound to ATPgS. | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7RLA
 
 | | Cryo-EM structure of human p97-R191Q mutant bound to ATPgS. | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
