6UO5
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 1 (CD1) Y363F mutant complexed with AR-42 | | Descriptor: | 1,2-ETHANEDIOL, AR-42, Histone deacetylase 6, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-10-14 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43934226 Å) | | Cite: | Structural Basis of Catalysis and Inhibition of HDAC6 CD1, the Enigmatic Catalytic Domain of Histone Deacetylase 6.
Biochemistry, 58, 2019
|
|
8TSM
 
 | |
7SNO
 
 | | Structure of Bacple_01701(H214N), a 6-O-galactose porphyran sulfatase | | Descriptor: | 1,2-ETHANEDIOL, 6-O-sulfo-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-6-O-sulfo-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, Arylsulfatase, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2021-10-28 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The porphyran degradation system of the human gut microbiota is complete, phylogenetically diverse and geographically structured across Asian populations
Biorxiv, 2023
|
|
2QEY
 
 | | Rat cytosolic PEPCK in complex with GTP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Sullivan, S.M, Holyoak, T. | | Deposit date: | 2007-06-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of rat cytosolic PEPCK: insight into the mechanism of phosphorylation and decarboxylation of oxaloacetic acid.
Biochemistry, 46, 2007
|
|
6QMR
 
 | | Complement factor D in complex with the inhibitor (S)-2-(2-((3'-(1-amino-2-hydroxyethyl)-[1,1'-biphenyl]-3-yl)methoxy)phenyl)acetic acid | | Descriptor: | 2-[2-[[3-[3-[(1~{S})-1-azanyl-2-oxidanyl-ethyl]phenyl]phenyl]methoxy]phenyl]ethanoic acid, Complement factor D | | Authors: | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
5GTP
 
 | |
9GNQ
 
 | | Microtubule-associated Kif5B IAK tail mutant bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J, Chegkazi, M.S, Steiner, R.A. | | Deposit date: | 2024-09-03 | | Release date: | 2024-11-20 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Microtubule association induces a Mg-free apo-like ADP pre-release conformation in kinesin-1 that is unaffected by its autoinhibitory tail.
Nat Commun, 16, 2025
|
|
5H36
 
 | | Crystal structures of the TRIC trimeric intracellular cation channel orthologue from Rhodobacter sphaeroides | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Uncharacterized protein TRIC | | Authors: | Kasuya, G, Hiraizumi, M, Hattori, M, Nureki, O. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.409 Å) | | Cite: | Crystal structures of the TRIC trimeric intracellular cation channel orthologues
Cell Res., 26, 2016
|
|
3JZ6
 
 | | Crystal structure of Mycobacterium smegmatis Branched Chain Aminotransferase in complex with pyridoxal-5'-phosphate at 1.9 angstrom. | | Descriptor: | Branched-chain amino acid aminotransferase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Castell, A, Mille, C, Unge, T. | | Deposit date: | 2009-09-23 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of mycobacterial branched-chain aminotransferase: implications for inhibitor design.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5MRH
 
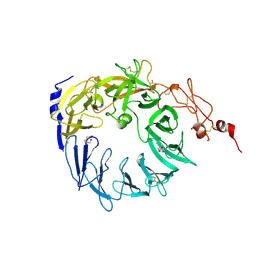 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with Triazolone 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(3-methylbutyl)-4~{H}-1,2,3-triazol-5-one, Sortilin, ... | | Authors: | Andersen, J.L, Strandbygaard, D, Thirup, S. | | Deposit date: | 2016-12-23 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The identification of novel acid isostere based inhibitors of the VPS10P family sorting receptor Sortilin.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
2QIS
 
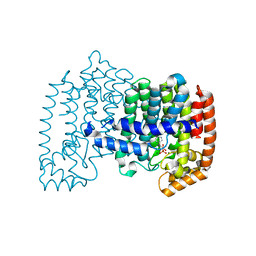 | | Crystal structure of human farnesyl pyrophosphate synthase T210S mutant bound to risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Kavanagh, K.L, Dunford, J.E, Hozjan, V, Evdokimov, A, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C.H, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-05 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human farnesyl pyrophosphate synthase T210S mutant bound to risedronate.
To be Published
|
|
6QOS
 
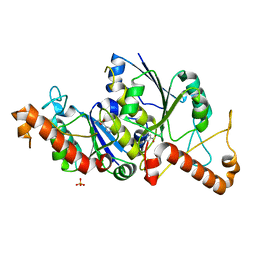 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 22 (Ethyl 1H-pyrazole-4-carboxylate) | | Descriptor: | SULFATE ION, ethyl 1~{H}-pyrazole-4-carboxylate, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6EYP
 
 | | X-ray structure of the unliganded uridine phosphorylase from Vibrio cholerae at 1.22A | | Descriptor: | GLYCEROL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Prokofev, I.I, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Lashkov, A.A. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-ray structure of the unliganded uridine phosphorylase from Vibrio cholerae at 1.22A
To Be Published
|
|
8IBC
 
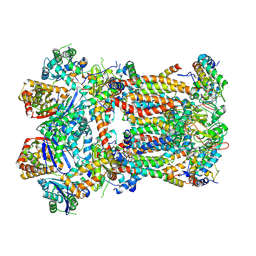 | | Respiratory complex CIII2, focus-refined of type IB, Wild type mouse under cold temperature | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, CARDIOLIPIN, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-02-10 | | Release date: | 2024-09-18 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 187, 2024
|
|
5GHW
 
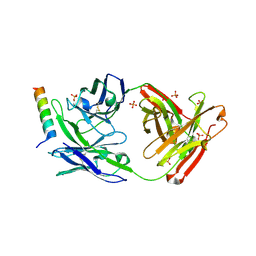 | | Crystal structure of broad neutralizing antibody 10E8 with long epitope bound | | Descriptor: | Endogenous retrovirus group K member 8 Env polyprotein, FAB 10E8 HEAVY CHAIN, FAB 10E8 LIGHT CHAIN, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Morante, K, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2016-06-21 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface
Sci Rep, 6, 2016
|
|
4ZSE
 
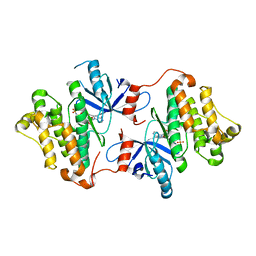 | |
9KUL
 
 | | Bovine Heart Cytochrome c Oxidase in the Xenon-bound Fully Oxidized State under Anaerobic Condition | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | Authors: | Muramoto, K, Shinzawa-Itoh, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The binding sites of carbon dioxide, nitrous oxide, and xenon reveal a putative exhaust channel for bovine cytochrome c oxidase.
J.Biol.Chem., 301, 2025
|
|
7G0V
 
 | | Crystal Structure of human FABP4 in complex with rac-(1R,2S)-2-[[3-(trifluoromethyl)phenoxy]methyl]cyclohexane-1-carboxylic acid, i.e. SMILES C1CC[C@H]([C@H](C1)C(=O)O)COc1cc(ccc1)C(F)(F)F with IC50=1.26466 microM | | Descriptor: | (1S,2R)-2-{[3-(trifluoromethyl)phenoxy]methyl}cyclohexane-1-carboxylic acid, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Buettelmann, B, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5D3Y
 
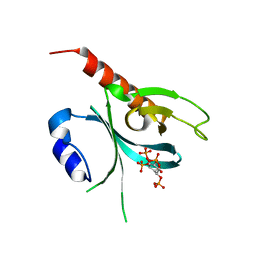 | | Crystal Structure of the P-Rex1 PH domain with Inositol-(1,3,4,5)-Tetrakisphosphate Bound | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2015-08-06 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3.
Structure, 24, 2016
|
|
9KUM
 
 | | Bovine Heart Cytochrome c Oxidase in the Xenon-bound Fully Reduced State | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | Authors: | Muramoto, K, Shinzawa-Itoh, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The binding sites of carbon dioxide, nitrous oxide, and xenon reveal a putative exhaust channel for bovine cytochrome c oxidase.
J.Biol.Chem., 301, 2025
|
|
7G0U
 
 | | Crystal Structure of human FABP4 in complex with 5-[(3-chlorophenyl)methyl]-6-hydroxy-1-methyl-4-morpholin-4-ylpyrimidin-2-one, i.e. SMILES N1=C(C(=C(N(C1=O)C)O)Cc1cc(Cl)ccc1)N1CCOCC1 with IC50=1.5 microM | | Descriptor: | 5-[(3-chlorophenyl)methyl]-6-hydroxy-1-methyl-4-(morpholin-4-yl)pyrimidin-2(1H)-one, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6UMF
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N-(5-{[1-(5-amino-1,3,4-thiadiazol-2-yl)piperidin-4-yl]oxy}-1,3,4-thiadiazol-2-yl)-2-phenylacetamide | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
6AYK
 
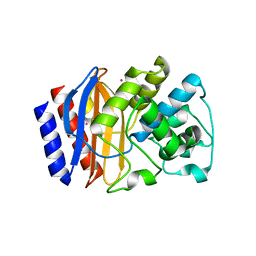 | |
7C6X
 
 | | Crystal structure of beta-glycosides-binding protein (W41A) of ABC transporter in an open state (Form I) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7FW7
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3-chloro-4-fluorophenyl)carbamoyl]cyclohexane-1-carboxylic acid, i.e. SMILES C(=O)([C@H]1[C@@H](C(=O)O)CCCC1)Nc1cc(c(cc1)F)Cl with IC50=12.0427 microM | | Descriptor: | (1S,2S)-2-[(3-chloro-4-fluorophenyl)carbamoyl]cyclohexane-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
