1VSH
 
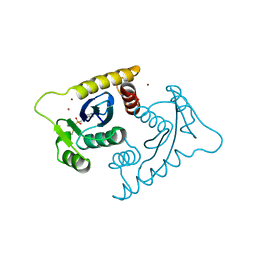 | | ASV INTEGRASE CORE DOMAIN WITH ZN(II) COFACTORS | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, INTEGRASE, ZINC ION | | Authors: | Bujacz, G, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1997-03-04 | | Release date: | 1997-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity.
J.Biol.Chem., 272, 1997
|
|
1VSI
 
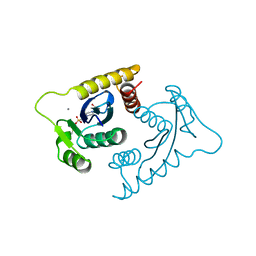 | | ASV INTEGRASE CORE DOMAIN WITH CA(II) COFACTOR | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, INTEGRASE | | Authors: | Bujacz, G, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1997-03-04 | | Release date: | 1997-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity.
J.Biol.Chem., 272, 1997
|
|
1VSJ
 
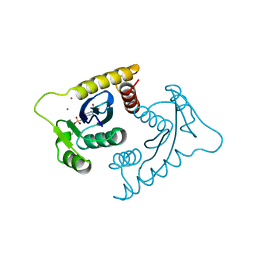 | | ASV INTEGRASE CORE DOMAIN WITH CD(II) COFACTORS | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, INTEGRASE | | Authors: | Bujacz, G, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1997-03-04 | | Release date: | 1997-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity.
J.Biol.Chem., 272, 1997
|
|
1VSK
 
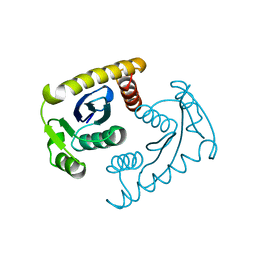 | |
1VSL
 
 | | ASV INTEGRASE CORE DOMAIN D64N MUTATION WITH ZINC CATION | | Descriptor: | PROTEIN (INTEGRASE), ZINC ION | | Authors: | Lubkowski, J, Yang, F, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1998-09-18 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for inactivating mutations and pH-dependent activity of avian sarcoma virus integrase.
J.Biol.Chem., 273, 1998
|
|
1VSM
 
 | |
1VSN
 
 | |
1VSO
 
 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex With the Antagonist (S)-ATPO at 1.85 A resolution | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, GLYCEROL, Glutamate receptor, ... | | Authors: | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-03-29 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
1VSQ
 
 | |
1VSR
 
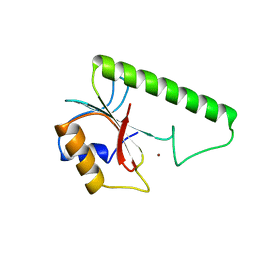 | | VERY SHORT PATCH REPAIR (VSR) ENDONUCLEASE FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (VSR ENDONUCLEASE), ZINC ION | | Authors: | Tsutakawa, S.E, Muto, T, Jingami, H, Kunishima, N, Ariyoshi, M, Kohda, D, Nakagawa, M, Morikawa, K. | | Deposit date: | 1999-02-13 | | Release date: | 1999-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and functional studies of very short patch repair endonuclease.
Mol.Cell, 3, 1999
|
|
1VST
 
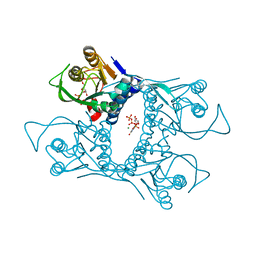 | | Symmetric Sulfolobus solfataricus uracil phosphoribosyltransferase with bound PRPP and GTP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kadziola, A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-09-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and kinetic studies of the allosteric transition in Sulfolobus solfataricus uracil phosphoribosyltransferase: Permanent activation by engineering of the C-terminus
J.Mol.Biol., 393, 2009
|
|
1VSU
 
 | |
1VSV
 
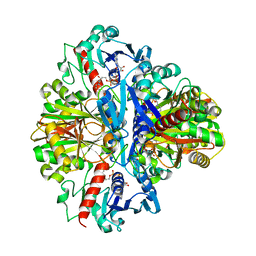 | |
1VT5
 
 | |
1VT6
 
 | |
1VT7
 
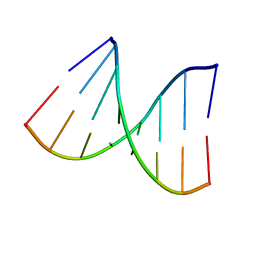 | |
1VT8
 
 | | Crystal structure of D(GGGCGCCC)-hexagonal form | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*C)-3') | | Authors: | Shakked, Z, Guerstein-Guzikevich, G, Eisenstein, M, Frolow, F, Rabinovich, D. | | Deposit date: | 1996-12-02 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The conformation of the DNA double helix in the crystal is dependent on its environment.
Nature, 342, 1989
|
|
1VT9
 
 | |
1VTA
 
 | |
1VTB
 
 | |
1VTC
 
 | |
1VTD
 
 | | UNUSUAL HELICAL PACKING IN CRYSTALS OF DNA BEARING A MUTATION HOT SPOT | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*GP*CP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*GP*CP*GP*CP*CP*GP*GP*T)-3') | | Authors: | Timsit, Y, Westhof, E, Fuchs, R.P.P, Moras, D. | | Deposit date: | 1996-12-12 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unusual helical packing in crystals of DNA bearing a mutation hot spot.
Nature, 341, 1989
|
|
1VTE
 
 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL A4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1990-05-21 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc. Natl. Acad. Sci. U.S.A., 87, 1990
|
|
1VTF
 
 | |
1VTG
 
 | | THE MOLECULAR STRUCTURE OF A DNA-TRIOSTIN A COMPLEX | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), triostin A | | Authors: | Wang, A.H.-J, Ughetto, G, Quigley, G.J, Hakoshima, T, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The molecular structure of a DNA-triostin A complex.
Science, 225, 1984
|
|
