1EKA
 
 | | NMR AND MOLECULAR MODELING REVEAL THAT DIFFERENT HYDROGEN BONDING PATTERNS ARE POSSIBLE FOR GU PAIRS: ONE HYDROGEN BOND FOR EACH GU PAIR IN R(GGCGUGCC)2 AND TWO FOR EACH GU PAIR IN R(GAGUGCUC)2 | | Descriptor: | RNA (5'-R(*GP*AP*GP*UP*GP*CP*UP*C)-3') | | Authors: | Chen, X, McDowell, J.A, Kierzek, R, Krugh, T.R, Turner, D.H. | | Deposit date: | 2000-03-07 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2).
Biochemistry, 39, 2000
|
|
2ZGL
 
 | |
3EOP
 
 | | Crystal Structure of the DUF55 domain of human thymocyte nuclear protein 1 | | Descriptor: | SULFATE ION, Thymocyte nuclear protein 1 | | Authors: | Yu, F, Song, A, Xu, C, Sun, L, Li, L, Tang, L, Hu, H, He, J. | | Deposit date: | 2008-09-29 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Determining the DUF55-domain structure of human thymocyte nuclear protein 1 from crystals partially twinned by tetartohedry
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3B3W
 
 | | Crystal structure of the S228A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine | | Descriptor: | Bacterial leucyl aminopeptidase, LEUCINE, SODIUM ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Zahniser, M.P.D, Milne, A, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3B69
 
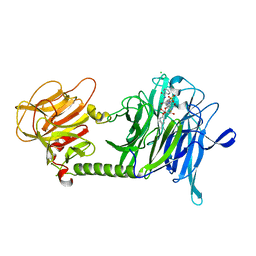 | | T cruzi Trans-sialidase complex with benzoylated NANA derivative | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-acetamido-9-(benzoylamino)-3,5,9-trideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CHLORIDE ION, ... | | Authors: | Buschiazzo, A. | | Deposit date: | 2007-10-28 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A new generation of specific Trypanosoma cruzi trans-sialidase inhibitors.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
3B7I
 
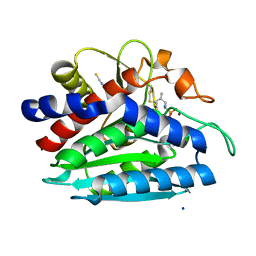 | | Crystal structure of the S228A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine phosphonic acid | | Descriptor: | Bacterial leucyl aminopeptidase, LEUCINE, LEUCINE PHOSPHONIC ACID, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Zahniser, M.P.D, Milne, A, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3H3V
 
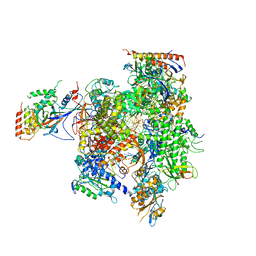 | | Yeast RNAP II containing poly(A)-signal sequence in the active site | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP*GP*CP*TP*GP*CP*TP*TP*TP*AP*TP*TP*GP*CP*AP*TP*T)-3', 5'-D(*CP*AP*GP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-R(*UP*GP*CP*AP*UP*UP*UP*CP*GP*CP*AP*AP*UP*AP*AP*A)-3', ... | | Authors: | Dengl, S, Cramer, P. | | Deposit date: | 2009-04-17 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Torpedo Nuclease Rat1 Is Insufficient to Terminate RNA Polymerase II in Vitro
J.Biol.Chem., 284, 2009
|
|
3CRU
 
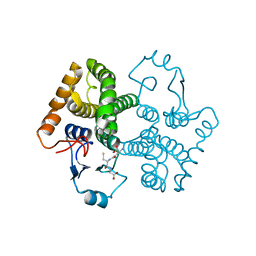 | | Structural characterization of an engineered allosteric protein | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Sagermann, M, Chapleau, R, DeLorimier, E, Lei, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Using affinity chromatography to engineer and characterize pH-dependent protein switches.
Protein Sci., 18, 2009
|
|
3CRT
 
 | | Structural characterization of an engineered allosteric protein | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Sagermann, M, Chapleau, R, DeLorimier, E, Lei, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Using affinity chromatography to engineer and characterize pH-dependent protein switches.
Protein Sci., 18, 2009
|
|
1EQH
 
 | | THE 2.7 ANGSTROM MODEL OF OVINE COX-1 COMPLEXED WITH FLURBIPROFEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, PROSTAGLANDIN H2 SYNTHASE-1, ... | | Authors: | Loll, P.J, Selinsky, B.S, Gupta, K, Sharkey, C.T. | | Deposit date: | 2000-04-04 | | Release date: | 2001-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of NSAID binding by prostaglandin H2 synthase: time-dependent and time-independent inhibitors elicit identical enzyme conformations.
Biochemistry, 40, 2001
|
|
3D4J
 
 | | Crystal structure of Human mevalonate diphosphate decarboxylase | | Descriptor: | Diphosphomevalonate decarboxylase, SULFATE ION | | Authors: | Voynova, N.E, Fu, Z, Battaile, K, Herdendorf, T.J, Kim, J.-J.P, Miziorko, H.M. | | Deposit date: | 2008-05-14 | | Release date: | 2008-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human mevalonate diphosphate decarboxylase: characterization, investigation of the mevalonate diphosphate binding site, and crystal structure.
Arch.Biochem.Biophys., 480, 2008
|
|
3CEV
 
 | | ARGINASE FROM BACILLUS CALDEVELOX, COMPLEXED WITH L-ARGININE | | Descriptor: | ARGININE, MANGANESE (II) ION, PROTEIN (ARGINASE) | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-15 | | Release date: | 1999-04-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
3CZJ
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE (N460T) IN COMPLEX WITH D-GALCTOPYRANOSYL-1-ONE | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Huber, R.E, Dugdale, M.L, Fraser, M.E, Tammam, S.D. | | Deposit date: | 2008-04-29 | | Release date: | 2009-04-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Practical Considerations When Using Temperature to Obtain Rate Constants and Activation Thermodynamics of Enzymes with Two Catalytic Steps: Native and N460T-beta-Galactosidase (E. coli) as Examples.
Protein J., 28, 2009
|
|
3D1Q
 
 | | Structure of the PTP-Like Phytase Expressed by Selenomonas Ruminantium at an Ionic Strength of 400 mM | | Descriptor: | CHLORIDE ION, GLYCEROL, Myo-inositol hexaphosphate phosphohydrolase | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2008-05-06 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effect of ionic strength and oxidation on the P-loop conformation of the protein tyrosine phosphatase-like phytase, PhyAsr.
Febs J., 275, 2008
|
|
1HT5
 
 | | THE 2.75 ANGSTROM RESOLUTION MODEL OF OVINE COX-1 COMPLEXED WITH METHYL ESTER FLURBIPROFEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN METHYL ESTER, PROSTAGLANDIN H2 SYNTHASE-1, ... | | Authors: | Selinsky, B.S, Gupta, K, Sharkey, C.T, Loll, P.J. | | Deposit date: | 2000-12-28 | | Release date: | 2001-04-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of NSAID binding by prostaglandin H2 synthase: time-dependent and time-independent inhibitors elicit identical enzyme conformations.
Biochemistry, 40, 2001
|
|
3CRW
 
 | | XPD_APO | | Descriptor: | HEXACYANOFERRATE(3-), XPD/Rad3 related DNA helicase | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Cell(Cambridge,Mass.), 133, 2008
|
|
1HTZ
 
 | | CRYSTAL STRUCTURE OF TEM52 BETA-LACTAMASE | | Descriptor: | BETA-LACTAMASE MUTANT TEM52 | | Authors: | Stevens, R.C, Orencia, M.C. | | Deposit date: | 2001-01-03 | | Release date: | 2001-03-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predicting the emergence of antibiotic resistance by directed evolution and structural analysis.
Nat.Struct.Biol., 8, 2001
|
|
3D1V
 
 | |
3D3H
 
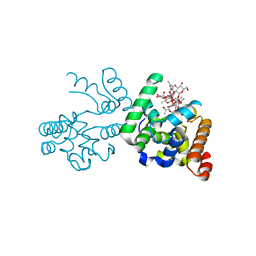 | | Crystal structure of a complex of the peptidoglycan glycosyltransferase domain from Aquifex aeolicus and neryl moenomycin A | | Descriptor: | (2R)-3-{[(S)-{[(2R,3R,4R,5S,6S)-3-{[(2S,3R,4R,5S,6R)-3-(acetylamino)-5-{[(2S,3R,4R,5S,6R)-3-(acetylamino)-5-{[(2R,3R,4S,5R,6S)-6-carbamoyl-3,4,5-trihydroxytetrahydro-2H-pyran-2-yl]oxy}-4-hydroxy-6-methyltetrahydro-2H-pyran-2-yl]oxy}-4-hydroxy-6-({[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]oxy}methyl)tetrahydro-2H-pyran-2-yl]oxy}-6-carbamoyl-4-(carbamoyloxy)-5-hydroxy-5-methyltetrahydro-2H-pyran-2-yl]oxy}(hydroxy)phosphoryl]oxy}-2-{[(2Z)-3,7-dimethylocta-2,6-dien-1-yl]oxy}propanoic acid, Penicillin-insensitive transglycosylase | | Authors: | Yuan, Y, Sliz, P, Walker, S. | | Deposit date: | 2008-05-11 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of the contacts anchoring moenomycin to peptidoglycan glycosyltransferases and implications for antibiotic design.
Acs Chem.Biol., 3, 2008
|
|
1IE0
 
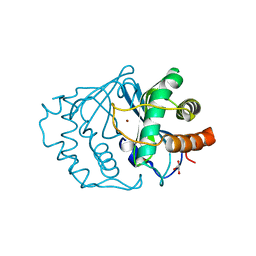 | | CRYSTAL STRUCTURE OF LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, GLYCEROL, ZINC ION | | Authors: | Hilgers, M.T, Ludwig, M.L. | | Deposit date: | 2001-04-05 | | Release date: | 2001-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the quorum-sensing protein LuxS reveals a catalytic metal site.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1ILZ
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI N156A ACTIVE SITE MUTANT pH 6.1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, OUTER MEMBRANE PHOSPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Van Eerde, J.H, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2001-05-09 | | Release date: | 2001-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn-His interaction in the catalytic triad.
Protein Sci., 10, 2001
|
|
1P6Y
 
 | | T4 LYSOZYME CORE REPACKING MUTANT M120Y/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-30 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Repacking the Core of T4 Lysozyme by Automated Design
J.Mol.Biol., 332, 2003
|
|
1OKB
 
 | | crystal structure of Uracil-DNA glycosylase from Atlantic cod (Gadus morhua) | | Descriptor: | CHLORIDE ION, GLYCEROL, URACIL-DNA GLYCOSYLASE | | Authors: | Leiros, I, Moe, E, Lanes, O, Smalas, A.O, Willassen, N.P. | | Deposit date: | 2003-07-21 | | Release date: | 2004-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Uracil-DNA Glycosylase from Atlantic Cod (Gadus Morhua) Reveals Cold-Adaptation Features
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P64
 
 | | T4 LYSOZYME CORE REPACKING MUTANT L133F/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-28 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
1PDC
 
 | |
