1BXM
 
 | | ENGINEERED BETA-CRYPTOGEIN COMPLEXED WITH ERGOSTEROL | | Descriptor: | BETA-CRYPTOGEIN, ERGOSTEROL | | Authors: | Boissy, G, Brunie, S. | | Deposit date: | 1998-10-05 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The 2.1 A structure of an elicitin-ergosterol complex: a recent addition to the Sterol Carrier Protein family.
Protein Sci., 8, 1999
|
|
1BXN
 
 | | THE CRYSTAL STRUCTURE OF RUBISCO FROM ALCALIGENES EUTROPHUS TO 2.7 ANGSTROMS. | | Descriptor: | PHOSPHATE ION, PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN), PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN) | | Authors: | Hansen, S, Vollan, V.B, Hough, E, Andersen, K. | | Deposit date: | 1998-10-06 | | Release date: | 1999-10-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of rubisco from Alcaligenes eutrophus reveals a novel central eight-stranded beta-barrel formed by beta-strands from four subunits.
J.Mol.Biol., 288, 1999
|
|
1BXO
 
 | | ACID PROTEINASE (PENICILLOPEPSIN) (E.C.3.4.23.20) COMPLEX WITH PHOSPHONATE INHIBITOR: METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUT YL] HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL) PHENYLPROPANOATE | | Descriptor: | GLYCEROL, METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUTYL]HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL)PHENYLPROPANOATE, PROTEIN (PENICILLOPEPSIN), ... | | Authors: | Khan, A.R, Parrish, J.C, Fraser, M.E, Smith, W.W, Bartlett, P.A, James, M.N.G. | | Deposit date: | 1998-10-07 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes.
Biochemistry, 37, 1998
|
|
1BXP
 
 | | SOLUTION NMR STRUCTURE OF THE COMPLEX OF ALPHA-BUNGAROTOXIN WITH A LIBRARY DERIVED PEPTIDE, 20 STRUCTURES | | Descriptor: | ALPHA-BUNGAROTOXIN, PEPTIDE MET-ARG-TYR-TYR-GLU-SER-SER-LEU-LYS-SER-TYR-PRO-ASP | | Authors: | Scherf, T, Balass, M, Fuchs, S, Katchalski-Katzir, E, Anglister, J. | | Deposit date: | 1998-08-23 | | Release date: | 1999-01-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the complex of alpha-bungarotoxin with a library-derived peptide.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1BXQ
 
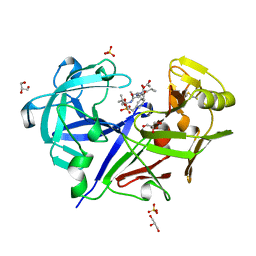 | | ACID PROTEINASE (PENICILLOPEPSIN) COMPLEX WITH PHOSPHONATE INHIBITOR. | | Descriptor: | 2-[(1R)-1-(N-(3-METHYLBUTANOYL)-L-VALYL-L-ASPARAGINYL)-AMINO)-3-METHYLBUTYL]HYDROXYPHOSPHINYLOXY]-3-PHENYLPROPANOIC ACID METHYLESTER, ACETATE ION, GLYCEROL, ... | | Authors: | Parrish, J.C, Khan, A.R, Fraser, M.E, Smith, W.W, Bartlett, P.A, James, M.N.G. | | Deposit date: | 1998-10-07 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes.
Biochemistry, 37, 1998
|
|
1BXR
 
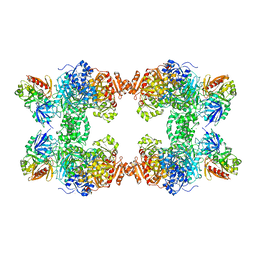 | | STRUCTURE OF CARBAMOYL PHOSPHATE SYNTHETASE COMPLEXED WITH THE ATP ANALOG AMPPNP | | Descriptor: | CARBAMOYL-PHOSPHATE SYNTHASE, CHLORIDE ION, L-ornithine, ... | | Authors: | Thoden, J.B, Wesenberg, G, Raushel, F.M, Holden, H.M. | | Deposit date: | 1998-10-08 | | Release date: | 1999-04-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carbamoyl phosphate synthetase: closure of the B-domain as a result of nucleotide binding.
Biochemistry, 38, 1999
|
|
1BXS
 
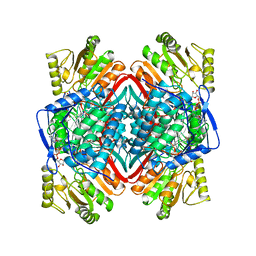 | | SHEEP LIVER CLASS 1 ALDEHYDE DEHYDROGENASE WITH NAD BOUND | | Descriptor: | ALDEHYDE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Moore, S.A, Baker, H.M, Blythe, T.J, Kitson, K.E, Kitson, T.M, Baker, E.N. | | Deposit date: | 1998-10-08 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases.
Structure, 6, 1998
|
|
1BXT
 
 | |
1BXU
 
 | | OXIDIZED PLASTOCYANIN FROM SYNECHOCOCCUS SP. | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Inoue, T, Sugawara, H, Hamanaka, S, Tsukui, H, Suzuki, E, Kohzuma, T, Kai, Y. | | Deposit date: | 1998-10-09 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure determinations of oxidized and reduced plastocyanin from the cyanobacterium Synechococcus sp. PCC 7942.
Biochemistry, 38, 1999
|
|
1BXV
 
 | | REDUCED PLASTOCYANIN FROM SYNECHOCOCCUS SP. | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Inoue, T, Sugawara, H, Hamanaka, S, Tsukui, H, Suzuki, E, Kohzuma, T, Kai, Y. | | Deposit date: | 1998-10-09 | | Release date: | 1999-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure determinations of oxidized and reduced plastocyanin from the cyanobacterium Synechococcus sp. PCC 7942.
Biochemistry, 38, 1999
|
|
1BXW
 
 | |
1BXX
 
 | |
1BXY
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L30 FROM THERMUS THERMOPHILUS AT 1.9 A RESOLUTION: CONFORMATIONAL FLEXIBILITY OF THE MOLECULE. | | Descriptor: | PROTEIN (RIBOSOMAL PROTEIN L30) | | Authors: | Fedorov, R, Nevskaya, N, Khairullina, A, Tishchenko, S, Mikhailov, A, Garber, M, Nikonov, S. | | Deposit date: | 1998-10-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ribosomal protein L30 from Thermus thermophilus at 1.9 A resolution: conformational flexibility of the molecule.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BXZ
 
 | | CRYSTAL STRUCTURE OF A THERMOPHILIC ALCOHOL DEHYDROGENASE SUBSTRATE COMPLEX FROM THERMOANAEROBACTER BROCKII | | Descriptor: | 2-BUTANOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, C, Heatwole, J, Soelaiman, S, Shoham, M. | | Deposit date: | 1998-10-09 | | Release date: | 2000-02-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of a thermophilic alcohol dehydrogenase substrate complex suggests determinants of substrate specificity and thermostability.
Proteins, 37, 1999
|
|
1BY0
 
 | |
1BY1
 
 | | DBL homology domain from beta-PIX | | Descriptor: | PROTEIN (PIX) | | Authors: | Aghazadeh, B, Zhu, K, Kubiseski, T.J, Liu, G.A, Pawson, T, Zheng, Y, Rosen, M.K. | | Deposit date: | 1998-10-22 | | Release date: | 1999-10-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and Mutagenesis of the Dbl Homology Domain
Nat.Struct.Biol., 5, 1998
|
|
1BY2
 
 | |
1BY3
 
 | | FHUA FROM E. COLI | | Descriptor: | N-OCTYL-2-HYDROXYETHYL SULFOXIDE, PROTEIN (FERRICHROME-IRON RECEPTOR PRECURSOR (FHUA)) | | Authors: | Locher, K.P, Rees, B, Koebnik, R, Mitschler, A, Moulinier, L, Rosenbusch, J.P, Moras, D. | | Deposit date: | 1998-10-22 | | Release date: | 1999-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes.
Cell(Cambridge,Mass.), 95, 1998
|
|
1BY4
 
 | | STRUCTURE AND MECHANISM OF THE HOMODIMERIC ASSEMBLY OF THE RXR ON DNA | | Descriptor: | DNA (5'-D(*C*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*A)-3'), PROTEIN (RETINOIC ACID RECEPTOR RXR-ALPHA), ... | | Authors: | Zhao, Q, Chasse, S.A, Devarakonda, S, Sierk, M.L, Ahvazi, B, Sigler, P.B, Rastinejad, F. | | Deposit date: | 1998-10-22 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of RXR-DNA interactions.
J.Mol.Biol., 296, 2000
|
|
1BY5
 
 | | FHUA FROM E. COLI, WITH ITS LIGAND FERRICHROME | | Descriptor: | FE (III) ION, FERRIC HYDROXAMATE UPTAKE PROTEIN, FERRICHROME, ... | | Authors: | Locher, K.P, Rees, B, Koebnik, R, Mitschler, A, Moulinier, L, Rosenbusch, J.P, Moras, D. | | Deposit date: | 1998-10-23 | | Release date: | 1999-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes.
Cell(Cambridge,Mass.), 95, 1998
|
|
1BY6
 
 | |
1BY7
 
 | |
1BY8
 
 | | THE CRYSTAL STRUCTURE OF HUMAN PROCATHEPSIN K | | Descriptor: | PROTEIN (PROCATHEPSIN K) | | Authors: | Lalonde, J.M, Zhao, B, Smith, W.W, Janson, C.A, Desjarlais, R.L, Tomaszek, T.A, Carr, T.J, Thompson, S.K, Yamashita, D.S, Veber, D.F, Abdel-Mequid, S.S. | | Deposit date: | 1998-10-27 | | Release date: | 1999-10-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of human procathepsin K.
Biochemistry, 38, 1999
|
|
1BY9
 
 | |
1BYA
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
