3EJK
 
 | |
5BUV
 
 | | X-ray structure of WbcA from Yersinia enterocolitica | | Descriptor: | 1,2-ETHANEDIOL, 6-AMINOPYRIMIDIN-2(1H)-ONE, PHOSPHATE ION, ... | | Authors: | Holden, H.M, Salinger, A.J, Brown, H.A, Thoden, J.B. | | Deposit date: | 2015-06-04 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical studies on WbcA, a sugar epimerase from Yersinia enterocolitica.
Protein Sci., 24, 2015
|
|
3RYK
 
 | | 1.63 Angstrom resolution crystal structure of dTDP-4-dehydrorhamnose 3,5-epimerase (rfbC) from Bacillus anthracis str. Ames with TDP and PPi bound | | Descriptor: | PYROPHOSPHATE 2-, THYMIDINE-5'-DIPHOSPHATE, dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Halavaty, A.S, Kuhn, M, Minasov, G, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-11 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme dTDP-4-dehydrorhamnose 3,5-epimerase (RfbC).
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6NDR
 
 | |
6C46
 
 | |
6DIN
 
 | | High resolutionstructure of apo dTDP-4-dehydrorhamnose 3,5-epimerase | | Descriptor: | SULFATE ION, dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Chang, C, Jedrzejczak, R, Chhor, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-23 | | Release date: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolutionstructure of apo dTDP-4-dehydrorhamnose 3,5-epimerase
To Be Published
|
|
7M15
 
 | | crystal structure of cj1430 in the presence of GDP-D-glycero-L-gluco-heptose, a GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase from campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-glycero-L-gluco-heptose, [(2R,3S,4R,5R)-5-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4R,5R,6S)-6-[(1R)-1,2-dihydroxyethyl]-3,4,5-trihydroxyoxan-2-yl dihydrogen diphosphate (non-preferred name) | | Authors: | Girardi, N.M, Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biosynthesis of d- glycero -l- gluco -Heptose in the Capsular Polysaccharides of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
7M14
 
 | | x-ray structure of cj1430 in the presence of GDP, a GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase from campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Girardi, N.M, Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biosynthesis of d- glycero -l- gluco -Heptose in the Capsular Polysaccharides of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
1WLT
 
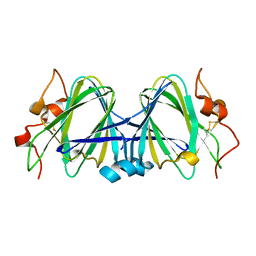 | | Crystal Structure of dTDP-4-dehydrorhamnose 3,5-epimerase homologue from Sulfolobus tokodaii | | Descriptor: | 176aa long hypothetical dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Watanabe, N, Sakai, N, Zilian, Z, Kawarabayasi, Y, Tanaka, I. | | Deposit date: | 2004-06-29 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of dTDP-4-dehydrorhamnose 3,5-epimerase homologue
to be published
|
|
2IXI
 
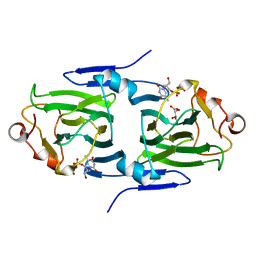 | | RmlC P aeruginosa with dTDP-xylose | | Descriptor: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, S,R MESO-TARTARIC ACID, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-08 | | Release date: | 2006-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rmlc, a C3' and C5' Carbohydrate Epimerase, Appears to Operate Via an Intermediate with an Unusual Twist Boat Conformation.
J.Mol.Biol., 365, 2007
|
|
2IXJ
 
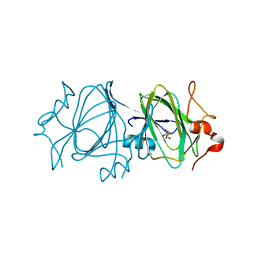 | | RmlC P aeruginosa native | | Descriptor: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, S,R MESO-TARTARIC ACID | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-08 | | Release date: | 2006-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Rmlc, a C3' and C5' Carbohydrate Epimerase, Appears to Operate Via an Intermediate with an Unusual Twist Boat Conformation.
J.Mol.Biol., 365, 2007
|
|
2IXH
 
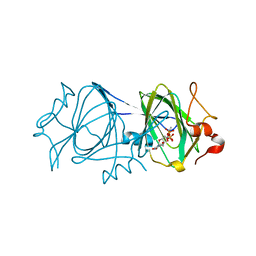 | | RmlC P aeruginosa with dTDP-rhamnose | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-08 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RmlC, a C3' and C5' carbohydrate epimerase, appears to operate via an intermediate with an unusual twist boat conformation.
J. Mol. Biol., 365, 2007
|
|
1RTV
 
 | | RmlC (dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase) crystal structure from Pseudomonas aeruginosa, apo structure | | Descriptor: | S,R MESO-TARTARIC ACID, dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Dong, C.J, Naismith, J.H. | | Deposit date: | 2003-12-10 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RmlC (dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase) crystal structure from Pseudomonas aeruginosa, apo structure
TO BE PUBLISHED
|
|
4HMZ
 
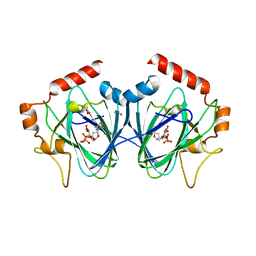 | | Crystal Structure of ChmJ, a 3'-monoepimerase from Streptomyces bikiniensis in complex with dTDP-quinovose | | Descriptor: | 1,2-ETHANEDIOL, Putative 3-epimerase in D-allose pathway, [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl (2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-methyltetrahydro-2H-pyran-2-yl dihydrogen diphosphate | | Authors: | Holden, H.M, Kubiak, R.L. | | Deposit date: | 2012-10-18 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Studies on a 3'-Epimerase Involved in the Biosynthesis of dTDP-6-deoxy-d-allose.
Biochemistry, 51, 2012
|
|
2B9U
 
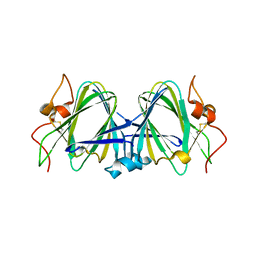 | | Crystal structure of dTDP-4-dehydrorhamnose 3,5-epimerase from sulfolobus tokodaii | | Descriptor: | hypothetical dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Rajakannan, V, Kondo, K, Mizushima, T, Suzuki, A, Yamane, T. | | Deposit date: | 2005-10-13 | | Release date: | 2006-10-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of dTDP-4-dehydrorhamnose 3,5-epimerase from sulfolobus tokodaii
TO BE PUBLISHED
|
|
1UPI
 
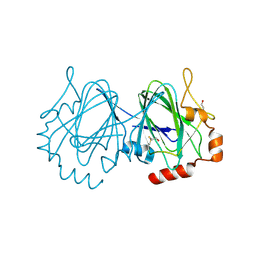 | | Mycobacterium tuberculosis rmlC epimerase (Rv3465) | | Descriptor: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE | | Authors: | Kim, C.-Y, Naranjo, C, Waldo, G.S, Lekin, T, Segelke, B.W, Kantardjieff, K.A, Zemla, A, Terwilliger, T, Rupp, B, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-10-05 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mycobacterium Tuberculosis Rmlc Epimerase (Rv3465): A Promising Drug-Target Structure in the Rhamnose Pathway
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4HN0
 
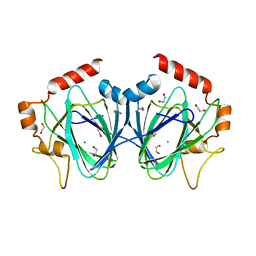 | |
4HN1
 
 | | Crystal Structure of H60N/Y130F double mutant of ChmJ, a 3'-monoepimerase from Streptomyces bikiniensis in complex with dTDP | | Descriptor: | 1,2-ETHANEDIOL, Putative 3-epimerase in D-allose pathway, THYMIDINE, ... | | Authors: | Holden, H.M, Kubiak, R.L. | | Deposit date: | 2012-10-18 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Studies on a 3'-Epimerase Involved in the Biosynthesis of dTDP-6-deoxy-d-allose.
Biochemistry, 51, 2012
|
|
1WA4
 
 | |
2C0Z
 
 | | The 1.6 A resolution crystal structure of NovW: a 4-keto-6-deoxy sugar epimerase from the novobiocin biosynthetic gene cluster of Streptomyces spheroides | | Descriptor: | 1,2-ETHANEDIOL, NOVW, SULFATE ION | | Authors: | Jakimowicz, P, Tello, M, Freel-Meyers, C.L, Walsh, C.T, Buttner, M.J, Field, R.A, Lawson, D.M. | | Deposit date: | 2005-09-09 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A Resolution Crystal Structure of Novw: A 4-Keto-6-Deoxy Sugar Epimerase from the Novobiocin Biosynthetic Gene Cluster of Streptomyces Spheroides
Proteins, 63, 2006
|
|
2IXL
 
 | | RmlC S. suis with dTDP-rhamnose | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, NICKEL (II) ION | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-08 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rmlc, a C3' and C5' Carbohydrate Epimerase, Appears to Operate Via an Intermediate with an Unusual Twist Boat Conformation.
J.Mol.Biol., 365, 2007
|
|
2IXC
 
 | | RmlC M. tuberculosis with dTDP-rhamnose | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE RMLC | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-07 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Rmlc, a C3' and C5' Carbohydrate Epimerase, Appears to Operate Via an Intermediate with an Unusual Twist Boat Conformation.
J.Mol.Biol., 365, 2007
|
|
2IXK
 
 | |
8DB5
 
 | | Crystal structure of the GDP-D-glycero-4-keto-d-lyxo-heptose-3,5-epimerase from Campylobacter jejuni, serotype HS:15 | | Descriptor: | CHLORIDE ION, GDP-D-glycero-4-keto-d-lyxo-heptose-3,5-epimerase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-14 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
8DAK
 
 | | Crystal structure of the GDP-D-glycero-4-keto-d-lyxo-heptose-3-epimerase from Campylobacter jejuni, serotype HS:3 | | Descriptor: | CHLORIDE ION, GDP-D-glycero-4-keto-d-lyxo-heptose-3-epimerase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-13 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
