6LQV
 
 | |
6LQT
 
 | |
6LQQ
 
 | |
6MDO
 
 | | The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Synaptosomal-associated protein 25, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
6LQP
 
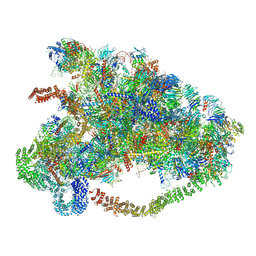 | |
6MDP
 
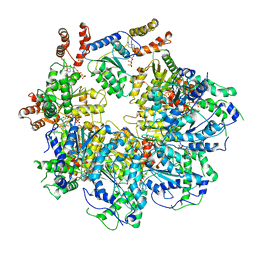 | | The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Synaptosomal-associated protein 25, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
6LQU
 
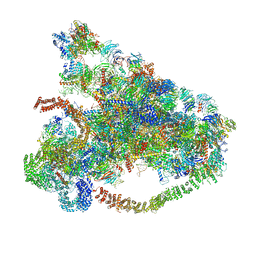 | |
6MAT
 
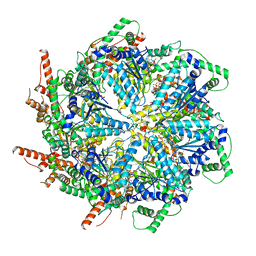 | | Cryo-EM structure of the essential ribosome assembly AAA-ATPase Rix7 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Rix7 mutant, unknown protein | | Authors: | Lo, Y.H, Sobhany, M, Hsu, A.L, Ford, B.L, Krahn, J.M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2018-08-28 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of the essential ribosome assembly AAA-ATPase Rix7.
Nat Commun, 10, 2019
|
|
6MDN
 
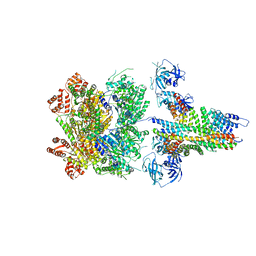 | | The 20S supercomplex engaging the SNAP-25 N-terminus (class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-soluble NSF attachment protein, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
6MDM
 
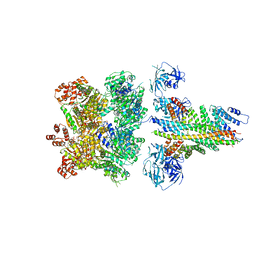 | | The 20S supercomplex engaging the SNAP-25 N-terminus (class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-soluble NSF attachment protein, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
6MCK
 
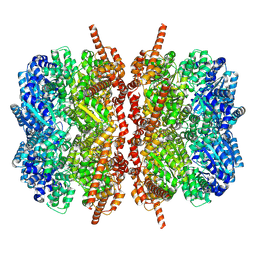 | | p97 D1D2 with CB5083 bound | | Descriptor: | 1-[4-(benzylamino)-7,8-dihydro-5H-pyrano[4,3-d]pyrimidin-2-yl]-2-methyl-1H-indole-4-carboxamide, Transitional endoplasmic reticulum ATPase | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2018-08-31 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Structural Basis of p97 Inhibition by the Site-Selective Anticancer Compound CB-5083.
Mol. Pharmacol., 95, 2019
|
|
6MSJ
 
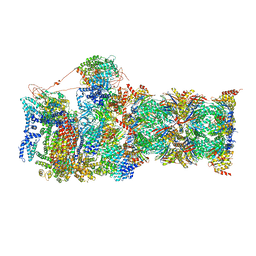 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSG
 
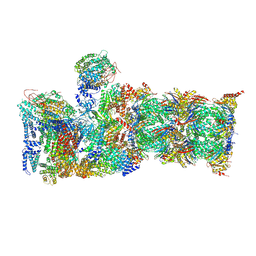 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSE
 
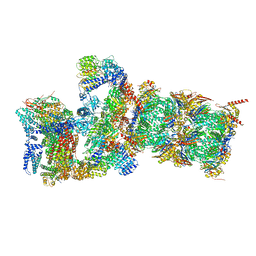 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6N8V
 
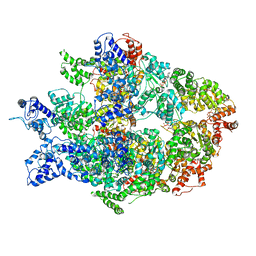 | | Hsp104DWB open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
6MSB
 
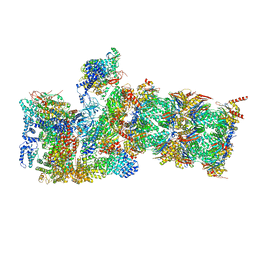 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSK
 
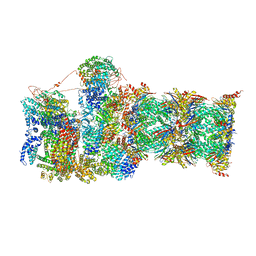 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSH
 
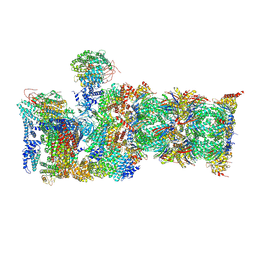 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6N8Z
 
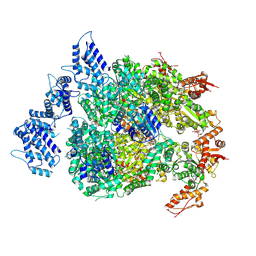 | | HSP104DWB extended conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2019-01-16 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
6MSD
 
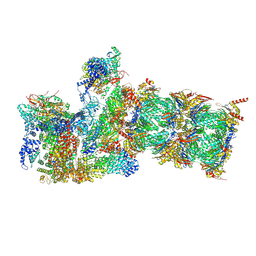 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6N8T
 
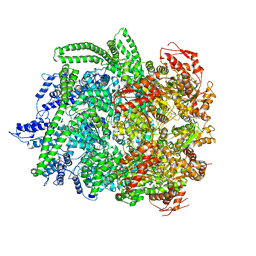 | | Hsp104DWB closed conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
6NDY
 
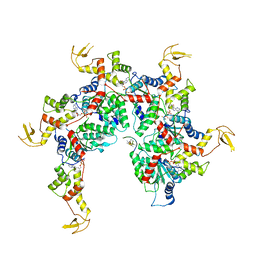 | | Vps4 with Cyclic Peptide Bound in the Central Pore | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Designed Cyclic Peptide, ... | | Authors: | Han, H, Fulcher, J.M, Dandey, V.P, Sundquist, W.I, Kay, M.S, Shen, P, Hill, C.P. | | Deposit date: | 2018-12-14 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases.
Elife, 8, 2019
|
|
6NYV
 
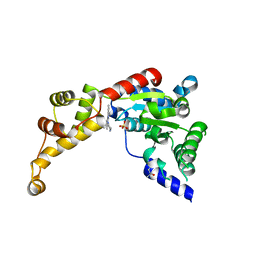 | | Structure of spastin AAA domain in complex with a quinazoline-based inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, N-(3-tert-butyl-1H-pyrazol-5-yl)-2-[(2R)-2-methylpiperazin-1-yl]quinazolin-4-amine, SULFATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-02-12 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.425 Å) | | Cite: | Designing Allele-Specific Inhibitors of Spastin, a Microtubule-Severing AAA Protein.
J. Am. Chem. Soc., 141, 2019
|
|
6NYY
 
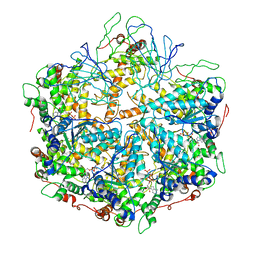 | | human m-AAA protease AFG3L2, substrate-bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AFG3-like protein 2, MAGNESIUM ION, ... | | Authors: | Lander, G.C, Puchades, C. | | Deposit date: | 2019-02-12 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unique Structural Features of the Mitochondrial AAA+ Protease AFG3L2 Reveal the Molecular Basis for Activity in Health and Disease.
Mol.Cell, 75, 2019
|
|
6NYW
 
 | | Structure of spastin AAA domain N527C mutant in complex with 8-fluoroquinazoline-based inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, N-(5-tert-butyl-1H-pyrazol-3-yl)-8-fluoro-2-[(2R)-2-methylpiperazin-1-yl]quinazolin-4-amine, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-02-12 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Designing Allele-Specific Inhibitors of Spastin, a Microtubule-Severing AAA Protein.
J. Am. Chem. Soc., 141, 2019
|
|
