5FML
 
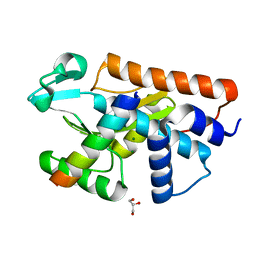 | | Crystal structure of the endonuclease from the PA subunit of influenza B virus bound to the PB2 subunit NLS peptide | | Descriptor: | GLYCEROL, MAGNESIUM ION, PA SUBUNIT OF INFLUENZA B POLYMERASE, ... | | Authors: | Guilligay, D, Gaudon, S, Cusack, S. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
6EUX
 
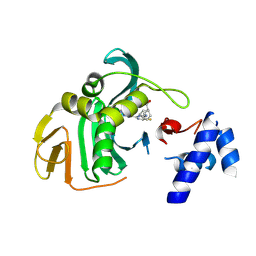 | |
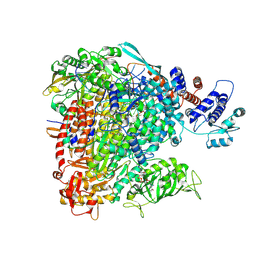
8BE0
 
 | |
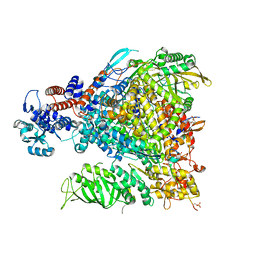
7Z42
 
 | |
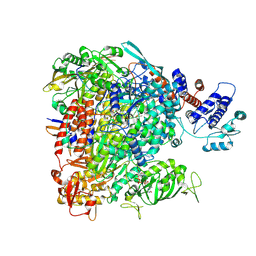
7R1F
 
 | |
4WRT
 
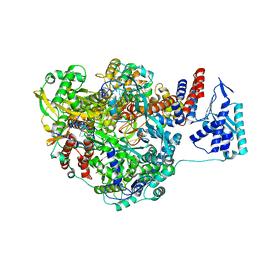 | | Crystal structure of Influenza B polymerase with bound vRNA promoter (form FluB2) | | Descriptor: | Influenza virus polymerase vRNA promoter 3' end, Influenza virus polymerase vRNA promoter 5' end, PA, ... | | Authors: | Reich, S, Guilligay, D, Pflug, A, Cusack, S. | | Deposit date: | 2014-10-25 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
8BDR
 
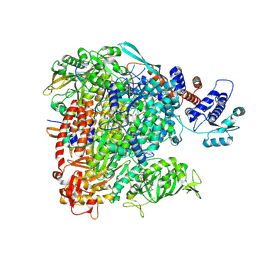 | |
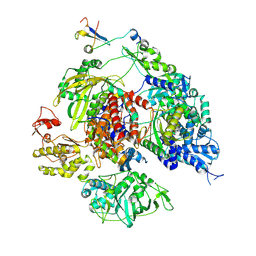
8RN3
 
 | |
8RN6
 
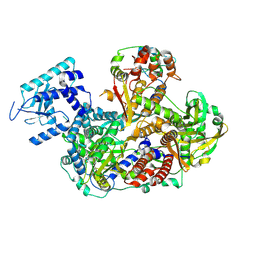 | |
8RN4
 
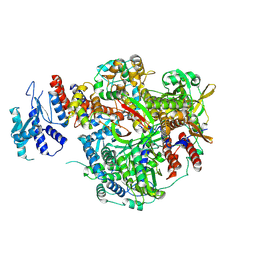 | |
8RN5
 
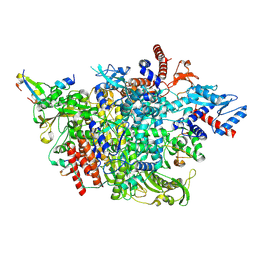 | |
6QCW
 
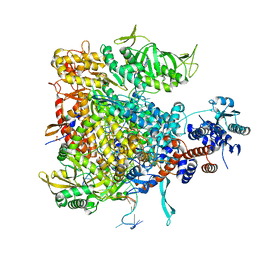 | |
8RN2
 
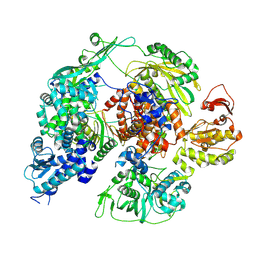 | | Monomeric apo-influenza B polymerase, encapsidase conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RN8
 
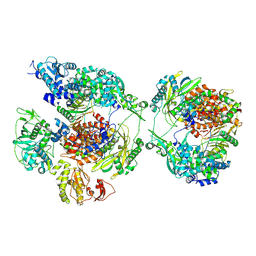 | | Influenza B polymerase pseudo-symmetrical apo-dimer (FluPol(E)|FluPol(S)) | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8BF5
 
 | |
6QCX
 
 | |
8RN7
 
 | |
6QCS
 
 | | Influenza B polymerase pre-initiation complex | | Descriptor: | 3 end, 5 end, Capped RNA, ... | | Authors: | Cusack, S, Kouba, T. | | Deposit date: | 2018-12-30 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural snapshots of actively transcribing influenza polymerase.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7Z43
 
 | |
6T0W
 
 | | Human Influenza B polymerase recycling complex | | Descriptor: | Polymerase PB2, Polymerase acidic protein, RNA-directed RNA polymerase catalytic subunit, ... | | Authors: | Wandzik, J.M, Kouba, T, Karuppasamy, M, Cusack, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
6QCT
 
 | | Influenza B polymerase elongation complex | | Descriptor: | 3 end, 5 end, MAGNESIUM ION, ... | | Authors: | Cusack, S, Kouba, T. | | Deposit date: | 2018-12-30 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural snapshots of actively transcribing influenza polymerase.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6QCV
 
 | |
8RNB
 
 | |
8RN9
 
 | |
4WSA
 
 | | Crystal structure of Influenza B polymerase bound to the vRNA promoter (FluB1 form) | | Descriptor: | Influenza B vRNA promoter 3' end, Influenza B vRNA promoter 5' end, PA, ... | | Authors: | Reich, S, Guilligay, D, Pflug, A, Cusack, S. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
