4UNM
 
 | |
5LQI
 
 | |
5LXZ
 
 | |
8GLV
 
 | |
7KZN
 
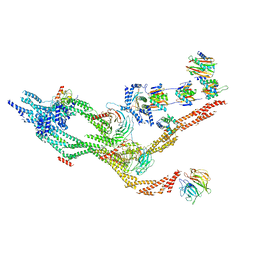 | | Outer dynein arm core subcomplex from C. reinhardtii | | Descriptor: | DC1, DC2, Dynein 11 kDa light chain, ... | | Authors: | Walton, T, Wu, H, Brown, A.B. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of a microtubule-bound axonemal dynein.
Nat Commun, 12, 2021
|
|
7KZM
 
 | | Outer dynein arm bound to doublet microtubules from C. reinhardtii | | Descriptor: | DC1, DC2, Dynein 11 kDa light chain, ... | | Authors: | Walton, T, Wu, H, Brown, A.B. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of a microtubule-bound axonemal dynein.
Nat Commun, 12, 2021
|
|
8VPQ
 
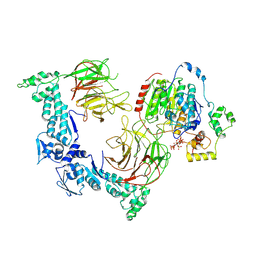 | | The structure of LSD1-CoREST-HDAC1 in complex with KBTBD4IPR310delinsTTYML | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Isoform 2 of Kelch repeat and BTB domain-containing protein 4, ... | | Authors: | Xie, X, Liau, B, Zheng, N. | | Deposit date: | 2024-01-16 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Converging mechanism of UM171 and KBTBD4 neomorphic cancer mutations.
Nature, 639, 2025
|
|
8VOJ
 
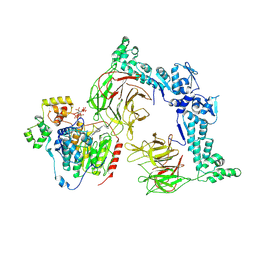 | | The Cryo-EM structure of LSD1-CoREST-HDAC1 in complex with KBTBD4 enhanced by UM171 and IP6 | | Descriptor: | (1r,4r)-N~1~-[(7P)-2-benzyl-7-(2-methyl-2H-tetrazol-5-yl)-9H-pyrimido[4,5-b]indol-4-yl]cyclohexane-1,4-diamine, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Xie, X, Mao, H, Liau, B, Zheng, N. | | Deposit date: | 2024-01-15 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Asymmetric Engagement of Dimeric CRL3KBTBD4 by the Molecular Glue UM171 Licenses Degradation of HDAC1/2 Complexes
To Be Published
|
|
8VRT
 
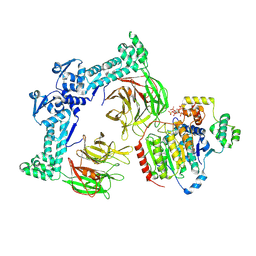 | | The structure of LSD1-CoREST-HDAC1 in complex with KBTBD4R313PRR mutant | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Kelch repeat and BTB domain-containing protein 4, ... | | Authors: | Xie, X, Liau, B, Zheng, N. | | Deposit date: | 2024-01-22 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Converging mechanism of UM171 and KBTBD4 neomorphic cancer mutations.
Nature, 639, 2025
|
|
9D1Y
 
 | |
9D1Z
 
 | |
9D1I
 
 | |
9DTG
 
 | | The cryo-EM structure of apo KBTBD4 | | Descriptor: | Isoform 2 of Kelch repeat and BTB domain-containing protein 4 | | Authors: | Xie, X, Mao, H, Liau, B, Zheng, N. | | Deposit date: | 2024-09-30 | | Release date: | 2024-11-27 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | UM171 glues asymmetric CRL3-HDAC1/2 assembly to degrade CoREST corepressors.
Nature, 639, 2025
|
|
9DTQ
 
 | | The structure of HDAC2-CoREST in complex with KBTBD4R313PRR mutant | | Descriptor: | Histone deacetylase 2, INOSITOL HEXAKISPHOSPHATE, Kelch repeat and BTB domain-containing protein 4, ... | | Authors: | Xie, X, Liau, B, Zheng, N. | | Deposit date: | 2024-10-01 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Converging mechanism of UM171 and KBTBD4 neomorphic cancer mutations.
Nature, 639, 2025
|
|
9GGL
 
 | | Cryo-EM structure of KBTBD4 WT-HDAC2 2:1 complex mediated by molecular glue UM171 | | Descriptor: | (1r,4r)-N~1~-[(7P)-2-benzyl-7-(2-methyl-2H-tetrazol-5-yl)-9H-pyrimido[4,5-b]indol-4-yl]cyclohexane-1,4-diamine, Histone deacetylase 2, Isoform 1 of Kelch repeat and BTB domain-containing protein 4, ... | | Authors: | Chen, Z, Chi, G, Pike, A.C.W, Montes, B, Bullock, A.N. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural mimicry of UM171 and neomorphic cancer mutants co-opts E3 ligase KBTBD4 for HDAC1/2 recruitment.
Nat Commun, 16, 2025
|
|
9GGN
 
 | | Cryo-EM structure of KBTBD4 WT-HDAC2 2:2 complex mediated by molecular glue UM171 | | Descriptor: | (1r,4r)-N~1~-[(7P)-2-benzyl-7-(2-methyl-2H-tetrazol-5-yl)-9H-pyrimido[4,5-b]indol-4-yl]cyclohexane-1,4-diamine, Histone deacetylase 2, Isoform 1 of Kelch repeat and BTB domain-containing protein 4, ... | | Authors: | Chen, Z, Chi, G, Pike, A.C.W, Montes, B, Bullock, A.N. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mimicry of UM171 and neomorphic cancer mutants co-opts E3 ligase KBTBD4 for HDAC1/2 recruitment.
Nat Commun, 16, 2025
|
|
9I2C
 
 | | Cryo-EM structure of KBTBD4 WT-HDAC2-CoREST1 2:1:1 complex mediated by molecular glue UM171 | | Descriptor: | (1r,4r)-N~1~-[(7P)-2-benzyl-7-(2-methyl-2H-tetrazol-5-yl)-9H-pyrimido[4,5-b]indol-4-yl]cyclohexane-1,4-diamine, Histone deacetylase 2, Isoform 1 of Kelch repeat and BTB domain-containing protein 4, ... | | Authors: | Chen, Z, Chi, G, Pike, A.C.W, Montes, B, Bullock, A.N. | | Deposit date: | 2025-01-20 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mimicry of UM171 and neomorphic cancer mutants co-opts E3 ligase KBTBD4 for HDAC1/2 recruitment.
Nat Commun, 16, 2025
|
|
9GGM
 
 | | Cryo-EM structure of KBTBD4 P313PRR mutant-HDAC2 2:2 complex | | Descriptor: | Histone deacetylase 2, Isoform 1 of Kelch repeat and BTB domain-containing protein 4, ZINC ION | | Authors: | Chen, Z, Chi, G, Pike, A.C.W, Montes, B, Bullock, A.N. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural mimicry of UM171 and neomorphic cancer mutants co-opts E3 ligase KBTBD4 for HDAC1/2 recruitment.
Nat Commun, 16, 2025
|
|
