6VJA
 
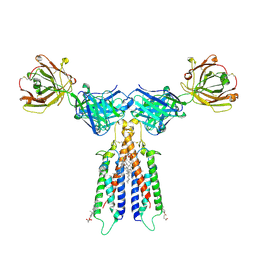 | | Structure of CD20 in complex with rituximab Fab | | Descriptor: | B-lymphocyte antigen CD20, CHOLESTEROL HEMISUCCINATE, Rituximab Fab heavy chain, ... | | Authors: | Rohou, A, Croll, T.I. | | Deposit date: | 2020-01-15 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of CD20 in complex with the therapeutic monoclonal antibody rituximab.
Science, 367, 2020
|
|
8VGO
 
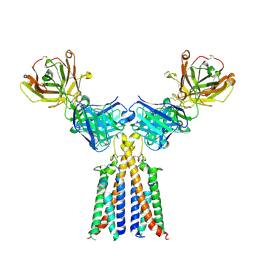 | | CryoEM structure of CD20 in complex with engineered conformationally rigid Rituximab.4DS Fab | | Descriptor: | B-lymphocyte antigen CD20, Rituximab.4DS Fab heavy chain, Rituximab.4DS Fab light chain | | Authors: | Kung, J.E, Jao, C.C, Arthur, C.P, Sudhamsu, J. | | Deposit date: | 2023-12-27 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Disulfide constrained Fabs overcome target size limitation for high-resolution single-particle cryo-EM.
Biorxiv, 2024
|
|
8VGN
 
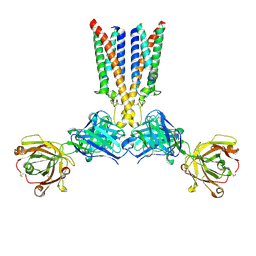 | | CryoEM structure of CD20 in complex with wild type Rituximab Fab | | Descriptor: | B-lymphocyte antigen CD20, Rituximab Fab heavy chain, Rituximab Fab light chain | | Authors: | Kung, J.E, Jao, C.C, Arthur, C.P, Sudhamsu, J. | | Deposit date: | 2023-12-27 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Disulfide constrained Fabs overcome target size limitation for high-resolution single-particle cryo-EM.
Biorxiv, 2024
|
|
6Y9A
 
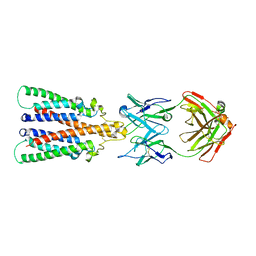 | |
6Y90
 
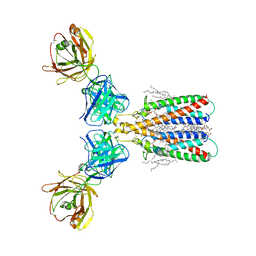 | | Structure of full-length CD20 in complex with Rituximab Fab | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, B-lymphocyte antigen CD20, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Kumar, A, Reyes, N. | | Deposit date: | 2020-03-06 | | Release date: | 2020-08-26 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Binding mechanisms of therapeutic antibodies to human CD20.
Science, 369, 2020
|
|
6Y97
 
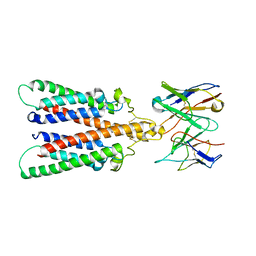 | |
6Y92
 
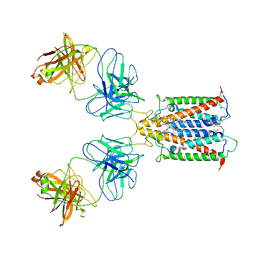 | |
8YVU
 
 | | structure of Ige receptor | | Descriptor: | CHOLESTEROL, High affinity immunoglobulin epsilon receptor subunit alpha, High affinity immunoglobulin epsilon receptor subunit beta, ... | | Authors: | Chen, M.Y, Su, Q, Shi, Y.G. | | Deposit date: | 2024-03-29 | | Release date: | 2024-11-06 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular mechanism of IgE-mediated Fc epsilon RI activation.
Nature, 637, 2025
|
|
9C3C
 
 | | Cryo-EM structure of native dystrophin-glycoprotein complex (DGC) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-dystroglycan, ... | | Authors: | Liu, S, Su, T, Xia, X, Zhou, Z.H. | | Deposit date: | 2024-05-31 | | Release date: | 2024-10-23 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Native DGC structure rationalizes muscular dystrophy-causing mutations.
Nature, 637, 2025
|
|
8K7T
 
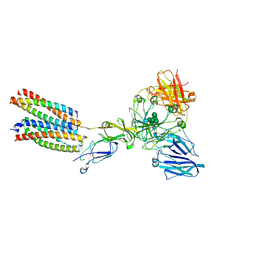 | | Mouse Fc epsilon RI in complex with mIgE Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, High affinity immunoglobulin epsilon receptor subunit beta, ... | | Authors: | Zhang, Z, Yui, M, Ohto, U, Shimizu, T. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Architecture of the high-affinity immunoglobulin E receptor.
Sci.Signal., 17, 2024
|
|
8Y84
 
 | | Structure of the high affinity receptor fc(epsilon)ri TM | | Descriptor: | CHOLESTEROL HEMISUCCINATE, High affinity immunoglobulin epsilon receptor subunit alpha, High affinity immunoglobulin epsilon receptor subunit beta, ... | | Authors: | Du, S, Deng, M.J, Xiao, J.Y. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-10 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into the high-affinity IgE receptor Fc epsilon RI complex.
Nature, 633, 2024
|
|
8Y81
 
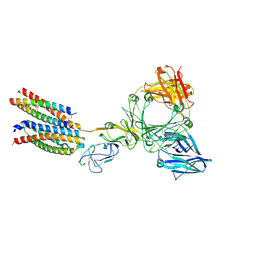 | | Structure of the ige-fc bound to its high affinity receptor fc(epsilon)ri | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Du, S, Deng, M.J, Xiao, J.Y. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into the high-affinity IgE receptor Fc epsilon RI complex.
Nature, 633, 2024
|
|
8YRJ
 
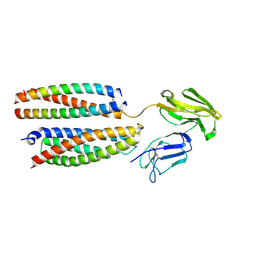 | | Mouse Fc epsilon RI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, High affinity immunoglobulin epsilon receptor subunit beta, ... | | Authors: | Zhang, Z, Yui, M, Ohto, U, Shimizu, T. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Architecture of the high-affinity immunoglobulin E receptor.
Sci.Signal., 17, 2024
|
|
8ZGS
 
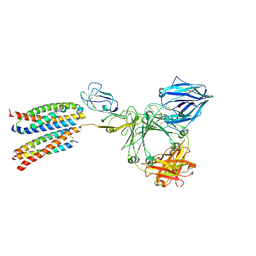 | | Structure of the ige-fc bound to its high affinity receptor fc(epsilon)ri state2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Du, S, Deng, M.J, Xiao, J.Y. | | Deposit date: | 2024-05-09 | | Release date: | 2024-07-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural insights into the high-affinity IgE receptor Fc epsilon RI complex.
Nature, 633, 2024
|
|
8ZGT
 
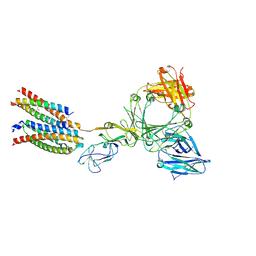 | | Structure of the ige-fc bound to its high affinity receptor fc(epsilon)ri state3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Du, S, Deng, M.J, Xiao, J.Y. | | Deposit date: | 2024-05-09 | | Release date: | 2024-07-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural insights into the high-affinity IgE receptor Fc epsilon RI complex.
Nature, 633, 2024
|
|
8YWA
 
 | | The structure of IgE receptor binding to IgE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, High affinity immunoglobulin epsilon receptor subunit beta, ... | | Authors: | Chen, M.Y, Su, Q, Shi, Y.G. | | Deposit date: | 2024-03-30 | | Release date: | 2024-11-06 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Molecular mechanism of IgE-mediated Fc epsilon RI activation.
Nature, 637, 2025
|
|
8YT8
 
 | | Cryo-EM structure of the dystrophin glycoprotein complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-sarcoglycan, ... | | Authors: | Wu, J.P, Yan, Z, Wan, L. | | Deposit date: | 2024-03-25 | | Release date: | 2024-12-11 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and assembly of the dystrophin glycoprotein complex.
Nature, 637, 2025
|
|
