7F5I
 
 | | X-ray structure of Clostridium perfringens-specific amidase endolysin | | Descriptor: | GLUTAMIC ACID, SODIUM ION, ZINC ION, ... | | Authors: | Kamitori, S, Tamai, E. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biochemical characterization of the Clostridium perfringens-specific Zn 2+ -dependent amidase endolysin, Psa, catalytic domain.
Biochem.Biophys.Res.Commun., 576, 2021
|
|
8C4D
 
 | | N-acetylmuramoyl-L-alanine amidase from Enterococcus faecium prophage genome | | Descriptor: | CHLORIDE ION, CITRATE ANION, N-acetylmuramoyl-L-alanine amidase, ... | | Authors: | Papageorgiou, A.C, Premetis, G.E, Labrou, N.E. | | Deposit date: | 2023-01-03 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and functional features of a broad-spectrum prophage-encoded enzybiotic from Enterococcus faecium.
Sci Rep, 13, 2023
|
|
6IDM
 
 | | Crystal structure of Peptidoglycan recognition protein (PGRP-S) with Tartaric acid at 3.20 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Bairagya, H.R, Shokeen, A, Sharma, P, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2018-09-10 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Peptidoglycan recognition protein (PGRP-S) with Tartaric acid at 3.20 A resolution
To Be Published
|
|
2L47
 
 | |
1J3G
 
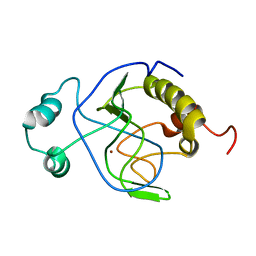 | | Solution structure of Citrobacter Freundii AmpD | | Descriptor: | AmpD protein, ZINC ION | | Authors: | Liepinsh, E, Genereux, C, Dehareng, D, Joris, B, Otting, G. | | Deposit date: | 2003-01-31 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Citrobacter freundii AmpD, Comparison with Bacteriophage T7 Lysozyme
and Homology with PGRP Domains
J.Mol.Biol., 327, 2003
|
|
6CKH
 
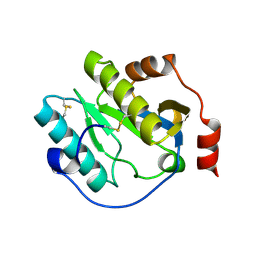 | | Manduca sexta Peptidoglycan Recognition Protein-1 | | Descriptor: | Peptidoglycan-recognition protein | | Authors: | Hu, Y. | | Deposit date: | 2018-02-28 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure and recognition mechanism of Manduca sexta peptidoglycan recognition protein-1.
Insect Biochem.Mol.Biol., 108, 2019
|
|
7DY5
 
 | | Structure of the ternary complex of peptidoglycan recognition protein-short (PGRP-S) with hexanoic acid and tartaric acid at 2.30A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Maurya, A, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-01-20 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the ternary complex of peptidoglycan recognition protein-short (PGRP-S)
To Be Published
|
|
1LBA
 
 | |
6FHG
 
 | |
1OHT
 
 | | Peptidoglycan recognition protein LB | | Descriptor: | 1,2-ETHANEDIOL, CG14704 PROTEIN, L(+)-TARTARIC ACID, ... | | Authors: | Kim, M.-S, Byun, M, Oh, B.-H. | | Deposit date: | 2003-05-31 | | Release date: | 2003-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Peptidoglycan Recognition Protein Lb from Drosophila Melanogaster
Nat.Immunol., 4, 2003
|
|
6SSC
 
 | | N-acetylmuramoyl-L-alanine amidase LysC from Clostridium intestinale URNW | | Descriptor: | GLYCEROL, N-acetylmuramoyl-L-alanine amidase, PHOSPHATE ION, ... | | Authors: | Hakansson, M, Al-Karadaghi, S, Kovacic, R, Plotka, M, Kaczorowska, A.K, Kaczorowski, T. | | Deposit date: | 2019-09-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Molecular Characterization of a Novel Lytic Enzyme LysC from Clostridium intestinale URNW and Its Antibacterial Activity Mediated by Positively Charged N -Terminal Extension.
Int J Mol Sci, 21, 2020
|
|
6SRT
 
 | | Endolysine N-acetylmuramoyl-L-alanine amidase LysCS from Clostridium intestinale URNW | | Descriptor: | GLYCEROL, N-acetylmuramoyl-L-alanine amidase, PHOSPHATE ION, ... | | Authors: | Hakansson, M, Al-Karadaghi, S, Plotka, M, Kaczorowska, A.-K, Kaczorowski, T. | | Deposit date: | 2019-09-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure and function of endolysines LysCS, LysC from Clostridium intestinale
To Be Published
|
|
6SU5
 
 | | Ph2119 endolysin from Thermus scotoductus MAT2119 bacteriophage Ph2119 | | Descriptor: | GLYCEROL, Lysozyme, PHOSPHATE ION, ... | | Authors: | Hakansson, M, Al-Karadaghi, S, Plotka, M, Kaczorowska, A.K, Kaczorowski, T. | | Deposit date: | 2019-09-13 | | Release date: | 2020-09-30 | | Last modified: | 2021-04-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Molecular Characterization of a Novel Lytic Enzyme LysC from Clostridium intestinale URNW and Its Antibacterial Activity Mediated by Positively Charged N -Terminal Extension.
Int J Mol Sci, 21, 2020
|
|
5XGY
 
 | | Crystal structure of peptidoglycan recognition protein (PGRP-S) at 2.45 A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Shokeen, A, Sharma, P, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-04-18 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of peptidoglycan recognition protein (PGRP-S) at 2.45 A resolution
To Be Published
|
|
5XZ4
 
 | | The X-tay structure of Bumblebee PGRP-SA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bumblebee peptidoglycan recognition protein SA, SULFATE ION | | Authors: | Liu, Y.J, Huang, J.X, Zhao, X.M, An, J.D. | | Deposit date: | 2017-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural Insights into the Preferential Binding of PGRP-SAs from Bumblebees and Honeybees to Dap-Type Peptidoglycans Rather than Lys-Type Peptidoglycans.
J Immunol., 202, 2019
|
|
4IVV
 
 | |
2R2K
 
 | | Crystal structure of the complex of camel peptidoglycan recognition protein with disaccharide at 3.2A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, L(+)-TARTARIC ACID, Peptidoglycan recognition protein | | Authors: | Sharma, P, Jain, R, Singh, N, Sharma, S, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-08-26 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of the complex of camel peptidoglycan recognition protein with disaccharide at 3.2A resolution
To be Published
|
|
2RKQ
 
 | |
2R90
 
 | | Crystal structure of cameline peptidoglycan recognition protein at 2.8A resolution | | Descriptor: | Peptidoglycan recognition protein | | Authors: | Sharma, P, Singh, N, Sinha, M, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-09-12 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of cameline peptidoglycan recognition protein at 2.8A resolution
To be Published
|
|
3LAT
 
 | |
4KNL
 
 | |
4KNK
 
 | | Crystal structure of Staphylococcus aureus hydrolase AmiA | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional autolysin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Buettner, F.M, Zoll, S, Stehle, T. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.124 Å) | | Cite: | Structure-function analysis of Staphylococcus aureus amidase reveals the determinants of peptidoglycan recognition and cleavage.
J.Biol.Chem., 289, 2014
|
|
6A89
 
 | | Crystal structure of the ternary complex of peptidoglycan recognition protein (PGRP-S) with Tartaric acid, Ribose and 2,6-DIAMINOPIMELIC ACID at 2.11 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2,6-DIAMINOPIMELIC ACID, GLYCEROL, ... | | Authors: | Bairagya, H.R, Shokeen, A, Sharma, P, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2018-07-06 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of the ternary complex of peptidoglycan recognition protein (PGRP-S) with Tartaric acid, Ribose and 2,6-DIAMINOPIMELIC ACID at 2.11 A resolution
To Be Published
|
|
3NG4
 
 | | Ternary complex of peptidoglycan recognition protein (PGRP-S) with Maltose and N-Acetylglucosamine at 1.7 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Multiligand specificity of pathogen-associated molecular pattern-binding site in peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
3NW3
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with the PGN Fragment at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, ALANINE, D-GLUTAMINE, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multiligand specificity of pathogen-associated molecular pattern-binding site in peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
