4TTC
 
 | |
4URP
 
 | | The Crystal structure of Nitroreductase from Saccharomyces cerevisiae | | Descriptor: | FATTY ACID REPRESSION MUTANT PROTEIN 2 | | Authors: | Song, H.-N, Woo, E.-J, Bang, S.-Y, Jung, D.-G, Park, S.-G. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Crystal Structure of the Fungal Nitroreductase Frm2 from Saccharomyces Cerevisiae.
Protein Sci., 24, 2015
|
|
8V5B
 
 | | Structure of the oxygen-insensitive NAD(P)H-dependent nitroreductase NfsB_Ec F70A/F108Y in complex with FMN | | Descriptor: | ACETATE ION, CHLORIDE ION, Dihydropteridine reductase, ... | | Authors: | Sharrock, A.V, Ackerley, D.F, Arcus, V. | | Deposit date: | 2023-11-30 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Evaluation of a Nitroreductase Engineered for Improved Activation of the 5-Nitroimidazole PET Probe SN33623.
Int J Mol Sci, 25, 2024
|
|
4TTB
 
 | |
6CZP
 
 | | 2.2 Angstrom Resolution Crystal Structure Oxygen-Insensitive NAD(P)H-dependent Nitroreductase NfsB from Vibrio vulnificus in Complex with FMN | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Kwon, K, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-09 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure Oxygen-Insensitive NAD(P)H-dependent Nitroreductase NfsB from Vibrio vulnificus in Complex with FMN.
To Be Published
|
|
5YAK
 
 | |
8OG3
 
 | | E. coli NfsB triple mutant T41L/N71S/F124T bound to citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Day, M.A, White, S.A, Hyde, E.I, Searle, P.F. | | Deposit date: | 2023-03-17 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
7JH4
 
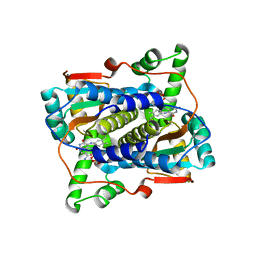 | | Crystal structure of NAD(P)H-flavin oxidoreductase (NfoR) from S. aureus complexed with reduced FMN and NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NAD(P)H-dependent oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zheng, Y, O'Neill, A.G, Beaupre, B.A, Liu, D, Moran, G.R. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NfoR: Chromate Reductase or Flavin Mononucleotide Reductase?
Appl.Environ.Microbiol., 86, 2020
|
|
7Q0O
 
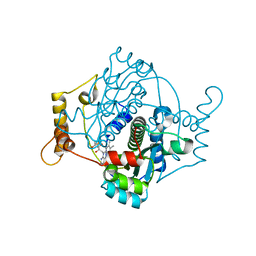 | | E. coli NfsA | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NADPH nitroreductase | | Authors: | White, S.A, Grainger, A, Parr, R, Day, M.A, Jarrom, D, Graziano, A, Searle, P.F, Hyde, E.I. | | Deposit date: | 2021-10-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | The 3D-structure, kinetics and dynamics of the E. coli nitroreductase NfsA with NADP + provide glimpses of its catalytic mechanism.
Febs Lett., 596, 2022
|
|
6WT2
 
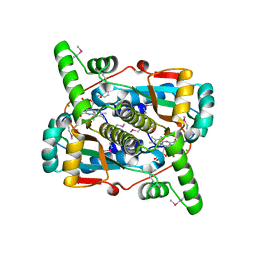 | | Crystal Structure of Putative NAD(P)H-Flavin Oxidoreductase from Neisseria meningitidis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-01 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and Structural Characterization of Diverse NfsB Chloramphenicol Reductase Enzymes from Human Pathogens.
Microbiol Spectr, 10, 2022
|
|
2R01
 
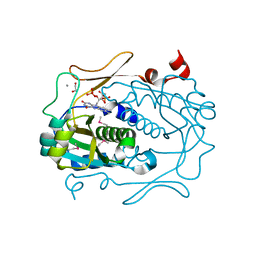 | |
5UU6
 
 | | The crystal structure of nitroreductase A from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Tan, K, Zhou, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of nitroreductase A from Vibrio parahaemolyticus RIMD 2210633
To Be Published
|
|
6TYK
 
 | |
1BKJ
 
 | | NADPH:FMN OXIDOREDUCTASE FROM VIBRIO HARVEYI | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH-FLAVIN OXIDOREDUCTASE, PHOSPHATE ION | | Authors: | Tanner, J.J, Lei, B, TU, S.-C, Krause, K.L. | | Deposit date: | 1998-07-08 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Flavin reductase P: structure of a dimeric enzyme that reduces flavin.
Biochemistry, 35, 1996
|
|
8DIL
 
 | | Crystal structure of putative nitroreductase from Salmonella enterica | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C, Skarina, T, Mesa, N, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-06-29 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative nitroreductase from Salmonella enterica
to be published
|
|
8DOR
 
 | |
8Q5G
 
 | |
3G14
 
 | |
3GH8
 
 | | Crystal structure of Mus musculus iodotyrosine deiodinase (IYD) bound to FMN and di-iodotyrosine (DIT) | | Descriptor: | 3,5-DIIODOTYROSINE, FLAVIN MONONUCLEOTIDE, Iodotyrosine dehalogenase 1, ... | | Authors: | Thomas, S.R, McTamney, P.M, Adler, J.M, LaRonde-LeBlanc, N, Rokita, S.E. | | Deposit date: | 2009-03-03 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of iodotyrosine deiodinase, a novel flavoprotein responsible for iodide salvage in thyroid glands.
J.Biol.Chem., 284, 2009
|
|
1F5V
 
 | | STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF A FLAVOPROTEIN FROM ESCHERICHIA COLI THAT REDUCES NITROCOMPOUNDS. ALTERATION OF PYRIDINE NUCLEOTIDE BINDING BY A SINGLE AMINO ACID SUBSTITUTION | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NADPH NITROREDUCTASE | | Authors: | Kobori, T, Sasaki, H, Lee, W.C, Zenno, S, Saigo, K, Murphy, M.E.P, Tanokura, M. | | Deposit date: | 2000-06-17 | | Release date: | 2001-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds: alteration of pyridine nucleotide binding by a single amino acid substitution.
J.Biol.Chem., 276, 2001
|
|
3PXV
 
 | |
2BKJ
 
 | |
4QLX
 
 | | Crystal structure of CLA-ER with product binding | | Descriptor: | 10-oxooctadecanoic acid, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Hou, F, Miyakawa, T, Tanokura, M. | | Deposit date: | 2014-06-13 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and reaction mechanism of a novel enone reductase.
Febs J., 282, 2015
|
|
2B67
 
 | | Crystal structure of the Nitroreductase Family Protein from Streptococcus pneumoniae TIGR4 | | Descriptor: | ACETIC ACID, COG0778: Nitroreductase, FLAVIN MONONUCLEOTIDE | | Authors: | Kim, Y, Volkart, L, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the Nitroreductase Family Protein from Streptococcus pneumoniae TIGR4
To be Published
|
|
1DS7
 
 | |
