4S21
 
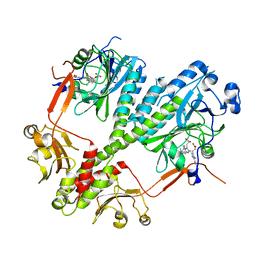 | | Crystal structure of the photosensory core module of bacteriophytochrome RPA3015 from R. palustris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome (Light-regulated signal transduction histidine kinase), PhyB1 | | Authors: | Yang, X, Stojkovi, E.A, Ozarowski, W.B, Moffat, K. | | Deposit date: | 2015-01-17 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Light Signaling Mechanism of Two Tandem Bacteriophytochromes.
Structure, 23, 2015
|
|
8BOR
 
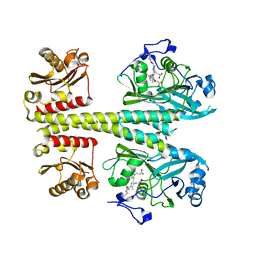 | | Photosensory module from DrBphP without PHY tongue | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Kurttila, M, Takala, H, Ihalainen, J.A. | | Deposit date: | 2022-11-15 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The interconnecting hairpin extension "arm": An essential allosteric element of phytochrome activity.
Structure, 31, 2023
|
|
7RZW
 
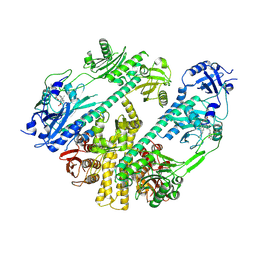 | | CryoEM structure of Arabidopsis thaliana phytochrome B | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B | | Authors: | Li, H, Burgie, E.S, Vierstra, R.D, Li, H. | | Deposit date: | 2021-08-27 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Plant phytochrome B is an asymmetric dimer with unique signalling potential.
Nature, 604, 2022
|
|
6TL4
 
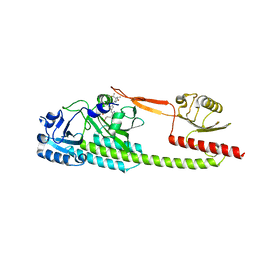 | | Photosensory module (PAS-GAF-PHY) of Glycine max phyB | | Descriptor: | PHYCOCYANOBILIN, Phytochrome | | Authors: | Nagano, S, Guan, K, Shenkutie, S.M, Hughes, J.E. | | Deposit date: | 2019-12-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into photoactivation and signalling in plant phytochromes.
Nat.Plants, 6, 2020
|
|
2VEA
 
 | |
8F5Z
 
 | | Composite map of CryoEM structure of Arabidopsis thaliana phytochrome A | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Li, H, Li, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The structure of Arabidopsis phytochrome A reveals topological and functional diversification among the plant photoreceptor isoforms.
Nat.Plants, 9, 2023
|
|
3C2W
 
 | |
8CO5
 
 | |
8YB4
 
 | | Pfr conformer of Arabidopsis thaliana phytochrome B in complex with phytochrome-interacting factor 6 | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, phytochrome B, phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 187, 2024
|
|
5UYR
 
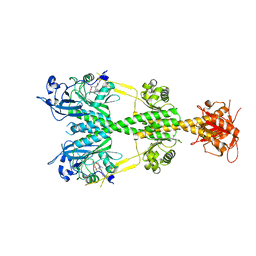 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP mutant D199A from Xanthomonas campestris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Klinke, S, Goldbaum, F.A, Bonomi, H.R. | | Deposit date: | 2017-02-24 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Pr-favoured variants of the bacteriophytochrome from the plant pathogen Xanthomonas campestris hint on light regulation of virulence-associated mechanisms.
Febs J., 288, 2021
|
|
6ET7
 
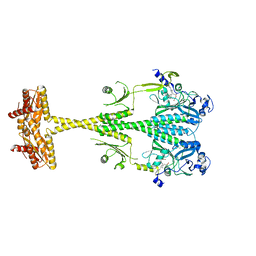 | |
3ZQ5
 
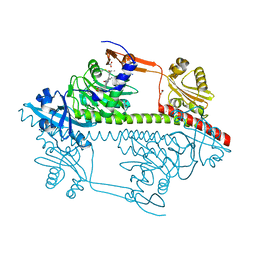 | | Structure of the Y263F mutant of the cyanobacterial phytochrome Cph1 | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Mailliet, J, Psakis, G, Sineshchekov, V, Essen, L.-O, Hughes, J. | | Deposit date: | 2011-06-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Spectroscopy and a High-Resolution Crystal Structure of Tyr263 Mutants of Cyanobacterial Phytochrome Cph1.
J.Mol.Biol., 413, 2011
|
|
9IUZ
 
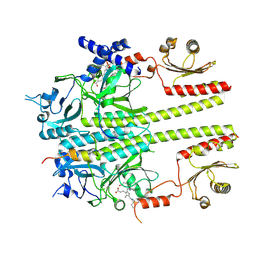 | | Constitutively active mutant(Y276H) of Arabidopsis phytochrome B(phyB) in complex with phytochrome-interacting factor 6(PIF6) | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B, Phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-07-22 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 187, 2024
|
|
9JRY
 
 | |
5AKP
 
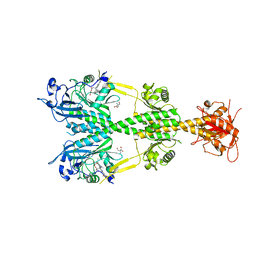 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP from Xanthomonas campestris bound to BV chromophore | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BILIVERDINE IX ALPHA, CHLORIDE ION, ... | | Authors: | Otero, L.H, Klinke, S, Goldbaum, F.A, Bonomi, H.R. | | Deposit date: | 2015-03-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the Full-Length Bacteriophytochrome from the Plant Pathogen Xanthomonas Campestris Provides Clues to its Long-Range Signaling Mechanism.
J.Mol.Biol., 428, 2016
|
|
5HSQ
 
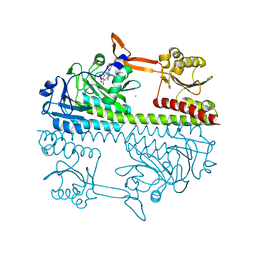 | | The surface engineered photosensory module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) in the Pr form, chromophore modelled with an endocyclic double bond in pyrrole ring A. | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome protein, CALCIUM ION, ... | | Authors: | Nagano, S, Scheerer, P, Zubow, K, Lamparter, T, Krauss, N. | | Deposit date: | 2016-01-26 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structures of the N-terminal Photosensory Core Module of Agrobacterium Phytochrome Agp1 as Parallel and Anti-parallel Dimers.
J.Biol.Chem., 291, 2016
|
|
3IBR
 
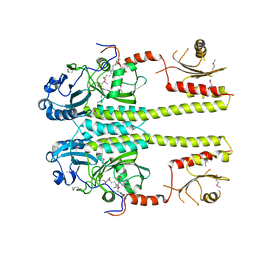 | |
6NDP
 
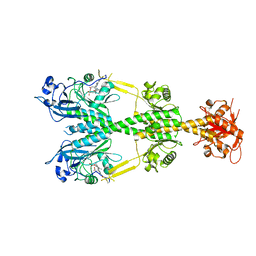 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP mutant L193Q from Xanthomonas campestris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Sirigu, S, Klinke, S, Rinaldi, J, Conforte, V, Malamud, F, Goldbaum, F.A, Chavas, L, Bonomi, H.R. | | Deposit date: | 2018-12-14 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Pr-favoured variants of the bacteriophytochrome from the plant pathogen Xanthomonas campestris hint on light regulation of virulence-associated mechanisms.
Febs J., 288, 2021
|
|
6NDO
 
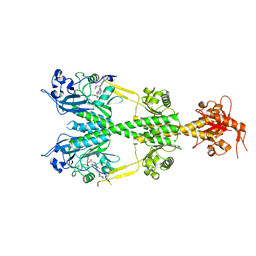 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP mutant L193N from Xanthomonas campestris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Sirigu, S, Klinke, S, Rinaldi, J, Conforte, V, Malamud, F, Goldbaum, F.A, Chavas, L, Bonomi, H.R. | | Deposit date: | 2018-12-14 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Pr-favoured variants of the bacteriophytochrome from the plant pathogen Xanthomonas campestris hint on light regulation of virulence-associated mechanisms.
Febs J., 288, 2021
|
|
5C5K
 
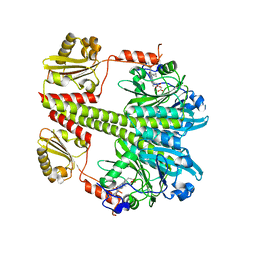 | | Structure of the Pfr form of a canonical phytochrome | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BILIVERDINE IX ALPHA, ... | | Authors: | Burgie, E.S, Vierstra, R.D. | | Deposit date: | 2015-06-20 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal Structure of Deinococcus Phytochrome in the Photoactivated State Reveals a Cascade of Structural Rearrangements during Photoconversion.
Structure, 24, 2016
|
|
3G6O
 
 | |
5I5L
 
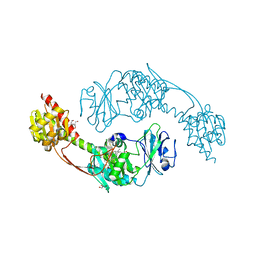 | | The photosensory module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) in the Pr form, chromophore modelled with an endocyclic double bond in pyrrole ring A | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome protein, CALCIUM ION, ... | | Authors: | Nagano, S, Scheerer, P, Zubow, K, Lamparter, T, Krauss, N. | | Deposit date: | 2016-02-15 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structures of the N-terminal Photosensory Core Module of Agrobacterium Phytochrome Agp1 as Parallel and Anti-parallel Dimers.
J.Biol.Chem., 291, 2016
|
|
9JLB
 
 | | Cryo-EM structure of phyB-PIF6beta complex | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B, Transcription factor PIF6 | | Authors: | Jia, H.L, Guan, Z.Y, Ding, J.Y, Wang, X.Y, Ma, L, Yin, P. | | Deposit date: | 2024-09-18 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insight into PIF6-mediated red light signal transduction of plant phytochrome B.
Cell Discov, 11, 2025
|
|
9IRK
 
 | | Cryo-EM structure of PhyB(Y276H,1-908)-PIF6beta complex | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B, Transcription factor PIF6 | | Authors: | Jia, H.L, Guan, Z.Y, Ding, J.Y, Wang, X.Y, Ma, L, Yin, P. | | Deposit date: | 2024-07-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight into PIF6-mediated red light signal transduction of plant phytochrome B.
Cell Discov, 11, 2025
|
|
9ITF
 
 | | Cryo-EM structure of full-length phyB(Y276H)-PIF6beta complex | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B, Transcription factor PIF6 | | Authors: | Jia, H.L, Guan, Z.Y, Ding, J.Y, Wang, X.Y, Ma, L, Yin, P. | | Deposit date: | 2024-07-20 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into PIF6-mediated red light signal transduction of plant phytochrome B.
Cell Discov, 11, 2025
|
|
