2LY8
 
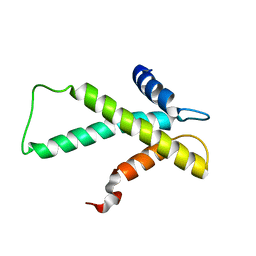 | | The budding yeast chaperone Scm3 recognizes the partially unfolded dimer of the centromere-specific Cse4/H4 histone variant | | Descriptor: | Budding yeast chaperone Scm3 | | Authors: | Hong, J, Feng, H, Zhou, Z, Ghirlando, R, Bai, Y. | | Deposit date: | 2012-09-13 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of Functionally Conserved Regions in the Structure of the Chaperone/CenH3/H4 Complex.
J.Mol.Biol., 425, 2013
|
|
7N8N
 
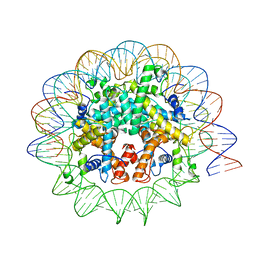 | | Melbournevirus nucleosome like particle | | Descriptor: | DNA (147-MER), Histone H2B-H2A doublet, Histone H4-H3 doublet | | Authors: | Liu, Y, Toner, C.M, Zhou, K, Bowerman, S, Luger, K. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Virus-encoded histone doublets are essential and form nucleosome-like structures.
Cell, 184, 2021
|
|
7LV8
 
 | | Structure of the Marseillevirus nucleosome | | Descriptor: | DNA (123-MER), Histone doublet Beta-Alpha (Alpha), Histone doublet Beta-Alpha (Beta), ... | | Authors: | Valencia-Sanchez, M.I, Abini-Agbomson, S, Armache, K.-J. | | Deposit date: | 2021-02-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of a virus-encoded nucleosome.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LV9
 
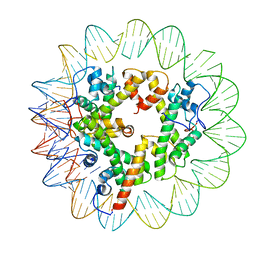 | | Marseillevirus heterotrimeric (hexameric) nucleosome | | Descriptor: | DNA (96-MER), Histone doublet Beta-Alpha (Alpha), Histone doublet Beta-Alpha (Beta), ... | | Authors: | Valencia-Sanchez, M.I, Abini-Agbomson, S, Armache, K.-J. | | Deposit date: | 2021-02-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The structure of a virus-encoded nucleosome.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6A7U
 
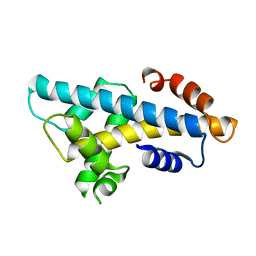 | | Crystal structure of histone H2A.Bbd-H2B dimer | | Descriptor: | Histone H2B type 2-E,Histone H2A-Bbd type 2/3 | | Authors: | Dai, L, Zhou, Z. | | Deposit date: | 2018-07-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the histone heterodimer containing histone variant H2A.Bbd.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
8UA7
 
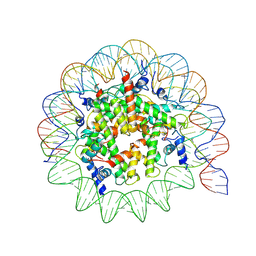 | | Medusavirus Nucleosome Core Particle | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Toner, C.M, Hoitsma, N.M, Weerawarana, S, Luger, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Characterization of Medusavirus encoded histones reveals nucleosome-like structures and a unique linker histone.
Nat Commun, 15, 2024
|
|
1EQZ
 
 | | X-RAY STRUCTURE OF THE NUCLEOSOME CORE PARTICLE AT 2.5 A RESOLUTION | | Descriptor: | 146 NUCLEOTIDES LONG DNA, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Hanson, B.L, Harp, J.M, Timm, D.E, Bunick, G.J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Asymmetries in the nucleosome core particle at 2.5 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2XQL
 
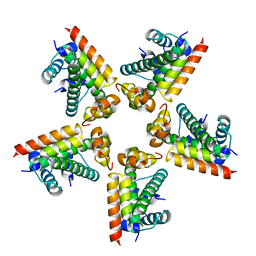 | | Fitting of the H2A-H2B histones in the electron microscopy map of the complex Nucleoplasmin:H2A-H2B histones (1:5). | | Descriptor: | HISTONE H2A-IV, HISTONE H2B 5 | | Authors: | Ramos, I, Martin-Benito, J, Finn, R, Bretana, L, Aloria, K, Arizmendi, J.M, Ausio, J, Muga, A, Valpuesta, J.M, Prado, A. | | Deposit date: | 2010-09-02 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (19.5 Å) | | Cite: | Nucleoplasmin Binds Histone H2A-H2B Dimers Through its Distal Face.
J.Biol.Chem., 285, 2010
|
|
1M1A
 
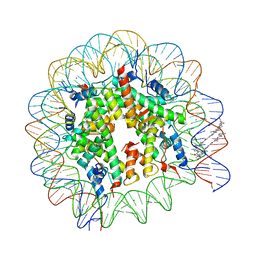 | | LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA | | Descriptor: | 3-AMINO-(DIMETHYLPROPYLAMINE), 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, 4-AMINO-(1-METHYLPYRROLE)-2-CARBOXYLIC ACID, ... | | Authors: | Suto, R.K, Edayathumangalam, R.S, White, C.L, Melander, C, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2002-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Nucleosome Core Particles in Complex with Minor Groove DNA-binding Ligands
J.MOL.BIOL., 326, 2003
|
|
6OM3
 
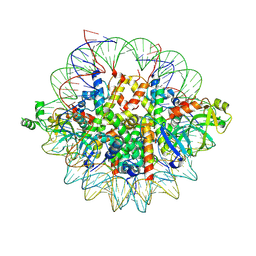 | | Crystal structure of the Orc1 BAH domain in complex with a nucleosome core particle | | Descriptor: | DNA (146-MER), DNA (147-MER), Histone H2A, ... | | Authors: | De Ioannes, P.E, Wang, M, Armache, K.-J. | | Deposit date: | 2019-04-18 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and function of the Orc1 BAH-nucleosome complex.
Nat Commun, 10, 2019
|
|
8QKT
 
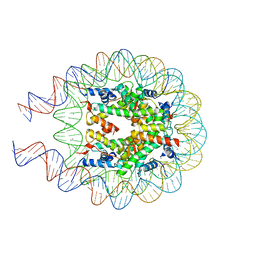 | |
6FQ8
 
 | | Class 3 : translocated nucleosome | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B, ... | | Authors: | Bilokapic, S, Halic, M. | | Deposit date: | 2018-02-13 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural rearrangements of the histone octamer translocate DNA.
Nat Commun, 9, 2018
|
|
3C9K
 
 | | Model of Histone Octamer Tubular Crystals | | Descriptor: | Histone H2A-IV, Histone H2B 7, Histone H3.2, ... | | Authors: | Frouws, T.D. | | Deposit date: | 2008-02-16 | | Release date: | 2009-01-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Histone octamer helical tubes suggest that an internucleosomal four-helix bundle stabilizes the chromatin fiber
Biophys.J., 96, 2009
|
|
1F66
 
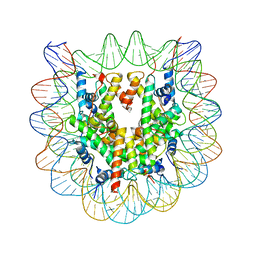 | | 2.6 A CRYSTAL STRUCTURE OF A NUCLEOSOME CORE PARTICLE CONTAINING THE VARIANT HISTONE H2A.Z | | Descriptor: | HISTONE H2A.Z, HISTONE H2B, HISTONE H3, ... | | Authors: | Suto, R.K, Clarkson, M.J, Tremethick, D.J, Luger, K. | | Deposit date: | 2000-06-20 | | Release date: | 2000-11-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a nucleosome core particle containing the variant histone H2A.Z.
Nat.Struct.Biol., 7, 2000
|
|
2L5A
 
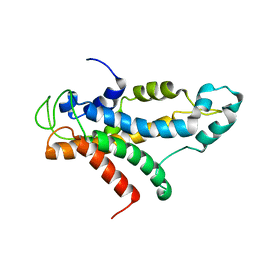 | | Structural basis for recognition of centromere specific histone H3 variant by nonhistone Scm3 | | Descriptor: | Histone H3-like centromeric protein CSE4, Protein SCM3, Histone H4 | | Authors: | Zhou, Z, Feng, H, Zhou, B, Ghirlando, R, Hu, K, Zwolak, A, Jenkins, L, Xiao, H, Tjandra, N, Wu, C, Bai, Y. | | Deposit date: | 2010-10-28 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3.
Nature, 472, 2011
|
|
2RVQ
 
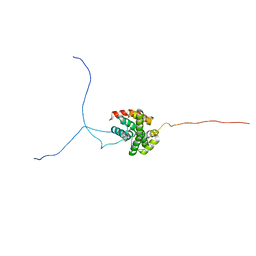 | | Solution structure of the isolated histone H2A-H2B heterodimer | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J | | Authors: | Moriwaki, Y, Yamane, T, Ohtomo, H, Ikeguchi, M, Kurita, J, Sato, M, Nagadoi, A, Shimojo, H, Nishimura, Y. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the isolated histone H2A-H2B heterodimer
Sci Rep, 6, 2016
|
|
8DK5
 
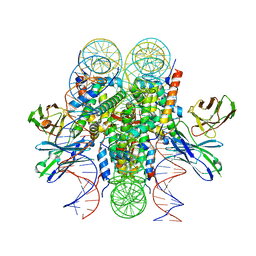 | | Structure of 187bp LIN28b nucleosome with site 0 mutation | | Descriptor: | DNA (187-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Lian, T, Guan, R, Bai, Y. | | Deposit date: | 2022-07-02 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
8DU4
 
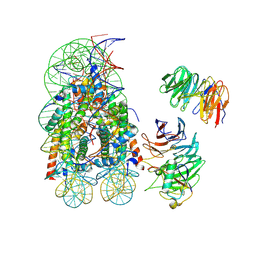 | | Complex between RbBP5-WDR5 and an H2B-ubiquitinated nucleosome | | Descriptor: | 601 DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Niklas, H.A, Rahman, S, Worden, E.J, Wolberger, C. | | Deposit date: | 2022-07-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Multistate structures of the MLL1-WRAD complex bound to H2B-ubiquitinated nucleosome.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3UTB
 
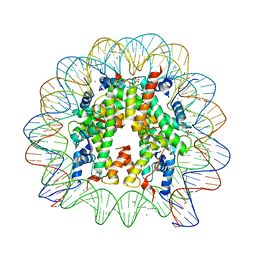 | | Crystal Structure of Nucleosome Core Particle Assembled with the 146b Alpha-Satellite Sequence (NCP146b) | | Descriptor: | 146-mer DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Chua, E.Y.D, Vasudevan, D, Davey, G.E, Wu, B, Davey, C.A. | | Deposit date: | 2011-11-25 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanics behind DNA sequence-dependent properties of the nucleosome
Nucleic Acids Res., 40, 2012
|
|
3LJA
 
 | |
3LEL
 
 | | Structural Insight into the Sequence-Dependence of Nucleosome Positioning | | Descriptor: | 147-MER DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Wu, B, Vasudevan, D, Davey, C.A. | | Deposit date: | 2010-01-15 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insight into the sequence dependence of nucleosome positioning
Structure, 18, 2010
|
|
3UT9
 
 | | Crystal Structure of Nucleosome Core Particle Assembled with a Palindromic Widom '601' Derivative (NCP-601L) | | Descriptor: | 145-mer DNA, CHLORIDE ION, Histone H2A, ... | | Authors: | Chua, E.Y.D, Vasudevan, D, Davey, G.E, Wu, B, Davey, C.A. | | Deposit date: | 2011-11-25 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanics behind DNA sequence-dependent properties of the nucleosome
Nucleic Acids Res., 40, 2012
|
|
4QLC
 
 | | Crystal structure of chromatosome at 3.5 angstrom resolution | | Descriptor: | CITRIC ACID, DNA (167-mer), H5, ... | | Authors: | Jiang, J.S, Zhou, B.R, Xiao, T.S, Bai, Y.W. | | Deposit date: | 2014-06-11 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structural Mechanisms of Nucleosome Recognition by Linker Histones.
Mol.Cell, 33 Suppl 1, 2015
|
|
1AOI
 
 | | COMPLEX BETWEEN NUCLEOSOME CORE PARTICLE (H3,H4,H2A,H2B) AND 146 BP LONG DNA FRAGMENT | | Descriptor: | HISTONE H2A, HISTONE H2B, HISTONE H3, ... | | Authors: | Luger, K, Maeder, A.W, Richmond, R.K, Sargent, D.F, Richmond, T.J. | | Deposit date: | 1997-07-03 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the nucleosome core particle at 2.8 A resolution.
Nature, 389, 1997
|
|
4R8P
 
 | | Crystal structure of the Ring1B/Bmi1/UbcH5c PRC1 ubiquitylation module bound to the nucleosome core particle | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase RING2, Ubiquitin-conjugating enzyme E2 D3, ... | | Authors: | McGinty, R.K, Henrici, R.C, Tan, S. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2846 Å) | | Cite: | Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome.
Nature, 514, 2014
|
|
