5DM3
 
 | | Crystal Structure of Glutamine Synthetase from Chromohalobacter salexigens DSM 3043(Csal_0679, TARGET EFI-550015) with bound ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-glutamine synthetase | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Glutamine Synthetase from Chromohalobacter salexigens DSM 3043(Csal_0679, TARGET EFI-550015) with bound ADP
To be published
|
|
2J9I
 
 | | Lengsin is a survivor of an ancient family of class I glutamine synthetases in eukaryotes that has undergone evolutionary re- engineering for a tissue-specific role in the vertebrate eye lens. | | Descriptor: | GLUTAMATE-AMMONIA LIGASE DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Wyatt, K, White, H.E, Wang, L, Bateman, O.A, Slingsby, C, Orlova, E.V, Wistow, G. | | Deposit date: | 2006-11-09 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Lengsin is a Survivor of an Ancient Family of Class I Glutamine Synthetases Re-Engineered by Evolution for a Role in the Vertebrate Lens.
Structure, 14, 2006
|
|
8WWU
 
 | | 1-naphthylamine GS in complex with AMP PNP | | Descriptor: | Glutamine synthetase, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Zhang, S.T, Zhou, N.Y. | | Deposit date: | 2023-10-26 | | Release date: | 2024-01-03 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of the 1-naphthylamine biodegradation pathway reveals a broad-substrate-spectrum enzyme catalyzing 1-naphthylamine glutamylation.
Elife, 13, 2024
|
|
8WWV
 
 | | 1-naphthylamine GS in complex with ADP and MetSox-P | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamine synthetase, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | Authors: | Zhang, S.T, Zhou, N.Y. | | Deposit date: | 2023-10-26 | | Release date: | 2024-01-03 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the 1-naphthylamine biodegradation pathway reveals a broad-substrate-spectrum enzyme catalyzing 1-naphthylamine glutamylation.
Elife, 13, 2024
|
|
7CQQ
 
 | | GmaS in complex with AMPPNP and MetSox | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQL
 
 | | Apo GmaS without ligand | | Descriptor: | Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQN
 
 | | GmaS in complex with AMPPCP | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQU
 
 | | GmaS/ADP/MetSox-P complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQX
 
 | | GmaS/ADP complex-Conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQW
 
 | | GmaS/ADP complex-Conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7V4H
 
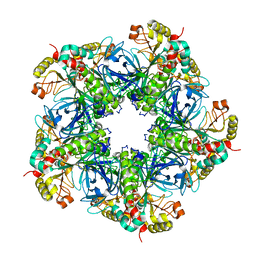 | |
8XLJ
 
 | |
4S17
 
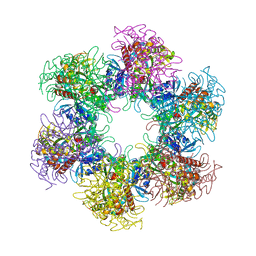 | | The crystal structure of glutamine synthetase from Bifidobacterium adolescentis ATCC 15703 | | Descriptor: | ACETATE ION, Glutamine synthetase, MAGNESIUM ION | | Authors: | Cuff, M, Tan, K, Mack, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-08 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of glutamine synthetase from Bifidobacterium adolescentis ATCC 15703
To be Published
|
|
8FBP
 
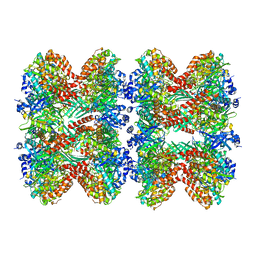 | | Glutamine synthetase from Pseudomonas aeruginosa, filament double-unit in compressed conformation | | Descriptor: | Glutamine synthetase | | Authors: | Phan, I.Q, Staker, B, Shek, R, Moser, T.H, Evans, J.E, van Voorhis, W.C, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Glutamine synthetase from Pseudomonas aeruginosa, filament double-unit in compressed conformation
To Be Published
|
|
1LGR
 
 | |
1HTO
 
 | |
8PVG
 
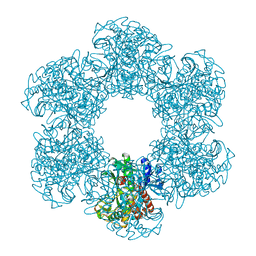 | | Structure of E. coli glutamine synthetase determined by cryoEM at 100 keV | | Descriptor: | Glutamine synthetase | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2WHI
 
 | | Crystal structure of Mycobacterium Tuberculosis Glutamine Synthetase in complex with a purine analogue inhibitor and L-methionine-S- sulfoximine phosphate. | | Descriptor: | 1-(3,4-dichlorobenzyl)-3,7-dimethyl-8-morpholin-4-yl-3,7-dihydro-1H-purine-2,6-dione, CHLORIDE ION, GLUTAMINE SYNTHETASE 1, ... | | Authors: | Nilsson, M.T, Krajewski, W.W, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2009-05-05 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterium Tuberculosis Glutamine Synthetase by Novel ATP-Competitive Inhibitors.
J.Mol.Biol., 393, 2009
|
|
4S0R
 
 | | Structure of GS-TnrA complex | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION, ... | | Authors: | Schumacher, M.A, Chinnam, N.G, Cuthbert, B, Tonthat, N.K. | | Deposit date: | 2015-01-04 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
9K2N
 
 | | Crystal structure of Glutamine Synthetase-apo | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glutamine synthetase, ... | | Authors: | Chandrasekaran, P, Killivalavan, A, Vaigundan, D, Yuvaraj, I, Manju, B, Sekar, K. | | Deposit date: | 2024-10-17 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of glutamine synthetase from ESKAPE pathogen Acinetobacter
baumannii
To Be Published
|
|
7EVT
 
 | | Crystal structure of the N-terminal degron-truncated human glutamine synthetase | | Descriptor: | Glutamine synthetase | | Authors: | Chek, M.F, Kim, S.Y, Mori, T, Hakoshima, T. | | Deposit date: | 2021-05-22 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of N-terminal degron-truncated human glutamine synthetase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
5LDF
 
 | |
9C28
 
 | |
4XYC
 
 | | NANOMOLAR INHIBITORS OF MYCOBACTERIUM TUBERCULOSIS GLUTAMINE SYNTHETASE 1: SYNTHESIS, BIOLOGICAL EVALUATION AND X-RAY CRYSTALLOGRAPHIC STUDIES | | Descriptor: | 9-phenyl-4H-imidazo[1,2-a]indeno[1,2-e]pyrazin-4-one, Glutamine synthetase 1 | | Authors: | Couturier, C, Silve, S, Morales, R, Ppessegue, B, Llopart, S, Nair, A, Bauer, A, Scheiper, B, poeverlein, c, Ganzhorn, A, Lagrange, S, Bacque, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Nanomolar inhibitors of Mycobacterium tuberculosis glutamine synthetase 1: Synthesis, biological evaluation and X-ray crystallographic studies.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8S59
 
 | |
