7WEZ
 
 | |
3IRW
 
 | | Structure of a c-di-GMP riboswitch from V. cholerae | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Smith, K.D. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding by a c-di-GMP riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4IUF
 
 | | Crystal Structure of Human TDP-43 RRM1 Domain in Complex with a Single-stranded DNA | | Descriptor: | 5'-D(*GP*TP*TP*GP*(XUA)P*GP*CP*GP*T)-3', TAR DNA-binding protein 43 | | Authors: | Kuo, P.H, Doudeva, L.G, Wang, Y.T, Yang, W.Z, Yuan, H.S. | | Deposit date: | 2013-01-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | The crystal structure of TDP-43 RRM1-DNA complex reveals the specific recognition for UG- and TG-rich nucleic acids.
Nucleic Acids Res., 42, 2014
|
|
4TU8
 
 | | STRUCTURE OF U2AF65 VARIANT WITH BRU5A6 DNA | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*DA*U)-3'), ... | | Authors: | MCLAUGHLIN, K.J, JENKINS, J.L, Agrawal, A.A, KIELKOPF, C.L. | | Deposit date: | 2014-06-24 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
9URH
 
 | |
4TU7
 
 | | Structure of U2AF65 D231V variant with BrU5 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*UP*U)-3'), GLYCEROL, ... | | Authors: | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2014-06-24 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2PE8
 
 | |
2OSQ
 
 | | NMR Structure of RRM-1 of Yeast NPL3 Protein | | Descriptor: | Nucleolar protein 3 | | Authors: | Deka, P, Bucheli, M, Skrisovska, L, Allain, F.H, Moore, C, Buratowski, S, Varani, G. | | Deposit date: | 2007-02-06 | | Release date: | 2007-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the yeast SR protein Npl3 and Interaction with mRNA 3'-end processing signals.
J.Mol.Biol., 375, 2008
|
|
4YUD
 
 | |
5D78
 
 | |
5DDQ
 
 | | L-glutamine riboswitch bound with L-glutamine soaked with Mn2+ | | Descriptor: | GLUTAMINE, L-glutamine riboswitch RNA (61-MER), MAGNESIUM ION, ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DDR
 
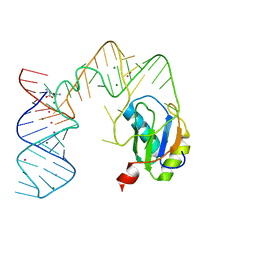 | | L-glutamine riboswitch bound with L-glutamine soaked with Cs+ | | Descriptor: | CESIUM ION, GLUTAMINE, L-glutamine riboswitch RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
4LMZ
 
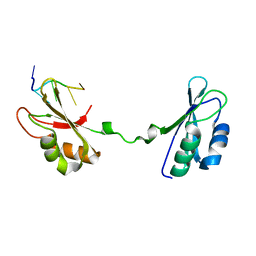 | |
4LJM
 
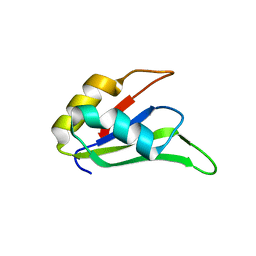 | |
1JMT
 
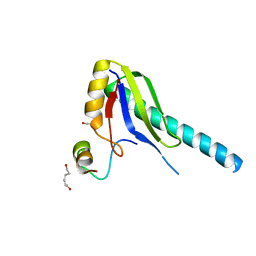 | | X-ray Structure of a Core U2AF65/U2AF35 Heterodimer | | Descriptor: | HEXANE-1,6-DIOL, SPLICING FACTOR U2AF 35 KDA SUBUNIT, SPLICING FACTOR U2AF 65 KDA SUBUNIT | | Authors: | Kielkopf, C.L, Rodionova, N.A, Green, M.R, Burley, S.K. | | Deposit date: | 2001-07-19 | | Release date: | 2001-09-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer.
Cell(Cambridge,Mass.), 106, 2001
|
|
1IQT
 
 | | Solution structure of the C-terminal RNA-binding domain of heterogeneous nuclear ribonucleoprotein D0 (AUF1) | | Descriptor: | heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Katahira, M, Miyanoiri, Y, Enokizono, Y, Matsuda, G, Nagata, T, Ishikawa, F, Uesugi, S. | | Deposit date: | 2001-08-01 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal RNA-binding domain of hnRNP D0 (AUF1), its interactions with RNA and DNA, and change in backbone dynamics upon complex formation with DNA.
J.Mol.Biol., 311, 2001
|
|
1L3K
 
 | | UP1, THE TWO RNA-RECOGNITION MOTIF DOMAIN OF HNRNP A1 | | Descriptor: | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN A1 | | Authors: | Vitali, J, Ding, J, Jiang, J, Zhang, Y, Krainer, A.R, Xu, R.-M. | | Deposit date: | 2002-02-27 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Correlated alternative side chain conformations in the RNA-recognition motif of heterogeneous nuclear ribonucleoprotein A1.
Nucleic Acids Res., 30, 2002
|
|
4BS2
 
 | | NMR structure of human TDP-43 tandem RRMs in complex with UG-rich RNA | | Descriptor: | 5'-R(*GP*UP*GP*UP*GP*AP*AP*UP*GP*AP*AP*UP)-3', TAR DNA-BINDING PROTEIN 43 | | Authors: | Lukavsky, P.J, Daujotyte, D, Tollervey, J.R, Ule, J, Stuani, C, Buratti, E, Baralle, F.E, Damberger, F.F, Allain, F.H.T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-11-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of Ug-Rich RNA Recognition by the Human Splicing Factor Tdp-43
Nat.Struct.Mol.Biol., 20, 2013
|
|
4C7Q
 
 | |
4W90
 
 | | Crystal structure of Bacillus subtilis cyclic-di-AMP riboswitch ydaO | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A, ... | | Authors: | Jones, C.P, Ferre-D'Amare, A.R. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.118 Å) | | Cite: | Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA.
Embo J., 33, 2014
|
|
6FQ1
 
 | |
4W92
 
 | | Crystal structure of Bacillus subtilis cyclic-di-AMP riboswitch ydaO | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, 1,2-ETHANEDIOL, C-di-AMP ribsoswitch, ... | | Authors: | Jones, C.P, Ferre-D'Amare, A.R. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA.
Embo J., 33, 2014
|
|
6GD2
 
 | | Structure of HuR RRM3 in complex with RNA | | Descriptor: | ELAV-like protein 1, RNA (5'-R(P*UP*UP*UP*AP*UP*UP*U)-3') | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-04-21 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
4UQT
 
 | | RRM-peptide structure in RES complex | | Descriptor: | PRE-MRNA-SPLICING FACTOR CWC26, U2 SNRNP COMPONENT IST3 | | Authors: | Tripsianes, K, Friberg, A, Barrandon, C, Seraphin, B, Sattler, M. | | Deposit date: | 2014-06-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Novel Protein-Protein Interaction in the Res (Retention and Splicing) Complex.
J.Biol.Chem., 289, 2014
|
|
9C7A
 
 | | Crystal structure of R149W neurodevelopmental disease-associated U2AF2 variant | | Descriptor: | DNA/RNA (5'-R(P*UP*U)-D(P*U)-R(P*UP*U)-D(P*(BRU))-R(P*CP*C)-3'), GLYCEROL, SODIUM ION, ... | | Authors: | Maji, D, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2024-06-10 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Recurrent Neurodevelopmentally Associated Variants of the Pre-mRNA Splicing Factor U2AF2 Alter RNA Binding Affinities and Interactions.
Biochemistry, 63, 2024
|
|
