5W0I
 
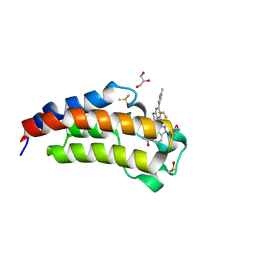 | | CREBBP Bromodomain in complex with Cpd8 (1-(3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydrofuran-3-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-[(3S)-oxolan-3-yl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-30 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
8CNB
 
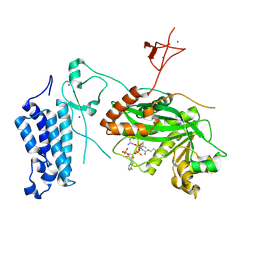 | | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
4I9O
 
 | | Crystal Structure of GACKIX L664C Tethered to 1-10 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[4-chloro-3-(trifluoromethyl)phenyl]-4-hydroxypiperidin-1-yl}-3-sulfanylpropan-1-one, CREB-binding protein | | Authors: | Wang, N, Meagher, J.L, Stuckey, J.A, Mapp, A.K. | | Deposit date: | 2012-12-05 | | Release date: | 2013-03-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ordering a dynamic protein via a small-molecule stabilizer.
J.Am.Chem.Soc., 135, 2013
|
|
7LVS
 
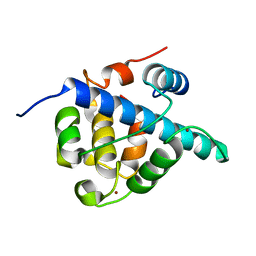 | | The CBP TAZ1 Domain in Complex with a CITED2-HIF-1-Alpha Fusion Peptide | | Descriptor: | Cbp/p300-interacting transactivator 2,Hypoxia-inducible factor 1-alpha, Histone lysine acetyltransferase CREBBP, ZINC ION | | Authors: | Appling, F.D, Berlow, R.B, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The molecular basis of allostery in a facilitated dissociation process.
Structure, 29, 2021
|
|
8CMZ
 
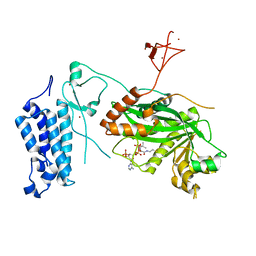 | | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-21 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
6DNQ
 
 | | HBZ77 in complex with KIX and c-Myb | | Descriptor: | 1,2-ETHANEDIOL, BZIP factor, CREB-binding protein, ... | | Authors: | Yang, K, Wright, P.E, Stanfield, R.L. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5U7G
 
 | | Crystal Structure of the Catalytic Core of CBP | | Descriptor: | CREB-binding protein, ZINC ION | | Authors: | Park, S, Stanfield, R.L, Martinez-Yamout, M.M, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Role of the CBP catalytic core in intramolecular SUMOylation and control of histone H3 acetylation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8CN0
 
 | | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-21 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
8CNA
 
 | | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA
To Be Published
|
|
8OG2
 
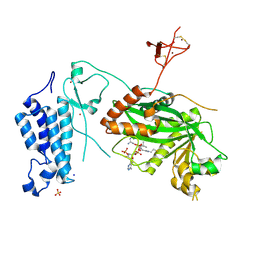 | | Crystal structure of CREBBP histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, SODIUM ION, SULFATE ION, ... | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-03-17 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of CREBBP histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
6DMX
 
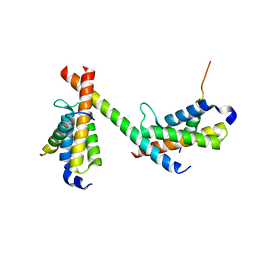 | | HBZ56 in complex with KIX and c-Myb | | Descriptor: | BZIP factor, CREB-binding protein, Transcriptional activator Myb | | Authors: | Yang, K, Wright, P.E, Stanfield, R.L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8CND
 
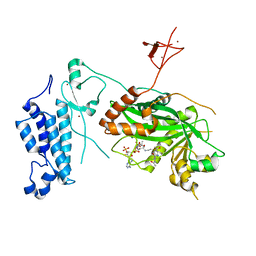 | | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA
To Be Published
|
|
2LQI
 
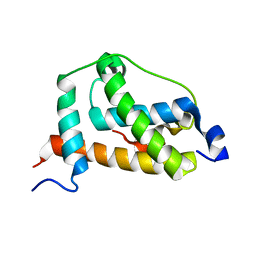 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2l3b conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2C52
 
 | | Structural diversity in CBP p160 complexes | | Descriptor: | CREB-BINDING PROTEIN, NUCLEAR RECEPTOR COACTIVATOR 1 | | Authors: | Waters, L.C, Yue, B, Veverka, V, Renshaw, P.S, Bramham, J, Matsuda, S, Frenkiel, T, Kelly, G, Muskett, F.W, Carr, M.D, Heery, D.M. | | Deposit date: | 2005-10-25 | | Release date: | 2006-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in p160/CREB-binding protein coactivator complexes.
J. Biol. Chem., 281, 2006
|
|
1R8U
 
 | | NMR structure of CBP TAZ1/CITED2 complex | | Descriptor: | CREB-binding protein, Cbp/p300-interacting transactivator 2, ZINC ION | | Authors: | De Guzman, R.N, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-10-28 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interaction of the TAZ1 domain of the CREB-binding protein with the activation domain of CITED2: regulation by competition between intrinsically unstructured ligands for non-identical binding sites.
J.Biol.Chem., 279, 2004
|
|
2LQH
 
 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2b3l conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5U4K
 
 | | NMR structure of the complex between the KIX domain of CBP and the transactivation domain 1 of p65 | | Descriptor: | CREB-binding protein, Transcription factor p65 | | Authors: | Lecoq, L, Raiola, L, Chabot, P.R, Cyr, N, Arseneault, G, Omichinski, J.G. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Nucleic Acids Res., 45, 2017
|
|
1TOT
 
 | | ZZ Domain of CBP- a Novel Fold for a Protein Interaction Module | | Descriptor: | CREB-binding protein, ZINC ION | | Authors: | Legge, G.B, Martinez-Yamout, M.A, Hambly, D.M, Trinh, T, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-06-15 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module
J.Mol.Biol., 343, 2004
|
|
1F81
 
 | | SOLUTION STRUCTURE OF THE TAZ2 DOMAIN OF THE TRANSCRIPTIONAL ADAPTOR PROTEIN CBP | | Descriptor: | CREB-BINDING PROTEIN, ZINC ION | | Authors: | De Guzman, R.N, Liu, H.L, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2000-06-28 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TAZ2 (CH3) domain of the transcriptional adaptor protein CBP.
J.Mol.Biol., 303, 2000
|
|
1KDX
 
 | | KIX DOMAIN OF MOUSE CBP (CREB BINDING PROTEIN) IN COMPLEX WITH PHOSPHORYLATED KINASE INDUCIBLE DOMAIN (PKID) OF RAT CREB (CYCLIC AMP RESPONSE ELEMENT BINDING PROTEIN), NMR 17 STRUCTURES | | Descriptor: | CBP, CREB | | Authors: | Radhakrishnan, I, Perez-Alvarado, G.C, Dyson, H.J, Wright, P.E. | | Deposit date: | 1997-09-16 | | Release date: | 1998-11-25 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions.
Cell(Cambridge,Mass.), 91, 1997
|
|
1JJS
 
 | | NMR Structure of IBiD, A Domain of CBP/p300 | | Descriptor: | CREB-BINDING PROTEIN | | Authors: | Lin, C.H, Hare, B.J, Wagner, G, Harrison, S.C, Maniatis, T, Fraenkel, E. | | Deposit date: | 2001-07-09 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A small domain of CBP/p300 binds diverse proteins: solution structure and functional studies.
Mol.Cell, 8, 2001
|
|
1KBH
 
 | | Mutual Synergistic Folding in the Interaction Between Nuclear Receptor Coactivators CBP and ACTR | | Descriptor: | CREB-BINDING PROTEIN, nuclear receptor coactivator | | Authors: | Demarest, S.J, Martinez-Yamout, M, Chung, J, Chen, H, Xu, W, Dyson, H.J, Evans, R.M, Wright, P.E. | | Deposit date: | 2001-11-06 | | Release date: | 2002-02-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators.
Nature, 415, 2002
|
|
5HP0
 
 | | Solution Structure of TAZ2-p53AD2 | | Descriptor: | CREB-binding protein,Cellular tumor antigen p53 fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HPD
 
 | | Solution Structure of TAZ2-p53TAD | | Descriptor: | CREB-binding protein,Cellular tumor antigen p53 fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-20 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2AGH
 
 | | Structural basis for cooperative transcription factor binding to the CBP coactivator | | Descriptor: | Crebbp protein, Myb proto-oncogene protein, Zinc finger protein HRX | | Authors: | De Guzman, R.N, Goto, N.K, Dyson, H.J, Wright, P.E. | | Deposit date: | 2005-07-26 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Cooperative Transcription Factor Binding to the CBP Coactivator
J.Mol.Biol., 355, 2006
|
|
