1RGI
 
 | | Crystal structure of gelsolin domains G1-G3 bound to actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Burtnick, L.D, Urosev, D, Irobi, E, Narayan, K, Robinson, R.C. | | Deposit date: | 2003-11-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the N-terminal half of gelsolin bound to actin: roles in severing, apoptosis and FAF
Embo J., 23, 2004
|
|
1RGJ
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN ALPHA-BUNGAROTOXIN AND MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR WITH ENHANCED ACTIVITY | | Descriptor: | MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR, long neurotoxin 1 | | Authors: | Bernini, A, Spiga, O, Ciutti, A, Scarselli, M, Bracci, L, Lozzi, L, Lelli, B, Neri, P, Niccolai, N. | | Deposit date: | 2003-11-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR and MD studies on the interaction between ligand peptides and alpha-bungarotoxin.
J.Mol.Biol., 339, 2004
|
|
1RGK
 
 | | RNASE T1 MUTANT GLU46GLN BINDS THE INHIBITORS 2'GMP AND 2'AMP AT THE 3' SUBSITE | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, CALCIUM ION, RIBONUCLEASE T1 | | Authors: | Granzin, J, Puras-Lutzke, R, Landt, O, Grunert, H.-P, Heinemann, U, Saenger, W, Hahn, U. | | Deposit date: | 1992-02-19 | | Release date: | 1993-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | RNase T1 mutant Glu46Gln binds the inhibitors 2'GMP and 2'AMP at the 3' subsite.
J.Mol.Biol., 225, 1992
|
|
1RGL
 
 | | RNASE T1 MUTANT GLU46GLN BINDS THE INHIBITORS 2'GMP AND 2'AMP AT THE 3' SUBSITE | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Granzin, J, Puras-Lutzke, R, Landt, O, Grunert, H.-P, Heinemann, U, Saenger, W, Hahn, U. | | Deposit date: | 1992-02-19 | | Release date: | 1993-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RNase T1 mutant Glu46Gln binds the inhibitors 2'GMP and 2'AMP at the 3' subsite.
J.Mol.Biol., 225, 1992
|
|
1RGN
 
 | | Structure of the reaction centre from Rhodobacter sphaeroides carotenoidless strain R-26.1 reconstituted with spheroidene | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Roszak, A.W, Frank, H.A, McKendrick, K, Mitchell, I.A, Cogdell, R.J, Isaacs, N.W. | | Deposit date: | 2003-11-12 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein Regulation of Carotenoid Binding: Gatekeeper and Locking Amino Acid Residues in Reaction Centers of Rhodobacter sphaeroides
STRUCTURE, 12, 2004
|
|
1RGO
 
 | | Structural Basis for Recognition of the mRNA Class II AU-Rich Element by the Tandem Zinc Finger Domain of TIS11d | | Descriptor: | Butyrate response factor 2, RNA (5'-R(*UP*UP*AP*UP*UP*UP*AP*UP*U)-3'), ZINC ION | | Authors: | Hudson, B.P, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-11-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d.
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
1RGP
 
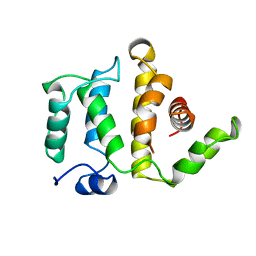 | | GTPASE-ACTIVATION DOMAIN FROM RHOGAP | | Descriptor: | RHOGAP | | Authors: | Barrett, T, Xiao, B, Dodson, E.J, Dodson, G, Ludbrook, S.B, Nurmahomed, K, Gamblin, S.J, Musacchio, A, Smerdon, S.J, Eccleston, J.F. | | Deposit date: | 1996-12-05 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the GTPase-activating domain from p50rhoGAP.
Nature, 385, 1997
|
|
1RGQ
 
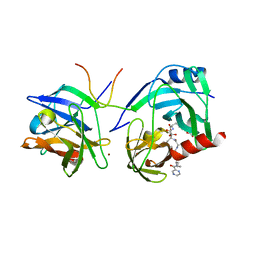 | | M9A HCV Protease complex with pentapeptide keto-amide inhibitor | | Descriptor: | N-(PYRAZIN-2-YLCARBONYL)LEUCYLISOLEUCYL-N~1~-{1-[2-({1-CARBOXY-2-[4-(PHOSPHONOOXY)PHENYL]ETHYL}AMINO)-1,1-DIHYDROXY-2-OXOETHYL]BUT-3-ENYL}-3-CYCLOHEXYLALANINAMIDE, NS3 Protease, NS4A peptide, ... | | Authors: | Liu, Y, Stoll, V.S, Richardson, P.L, Saldivar, A, Klaus, J.L, Molla, A, Kohlbrenner, W, Kati, W.M. | | Deposit date: | 2003-11-12 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Hepatitis C NS3 protease inhibition by peptidyl-alpha-ketoamide inhibitors: kinetic mechanism and structure.
Arch.Biochem.Biophys., 421, 2004
|
|
1RGR
 
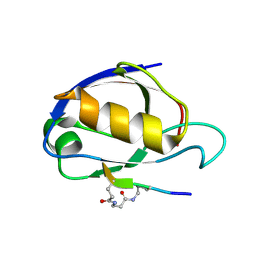 | | Cyclic Peptides Targeting PDZ Domains of PSD-95: Structural Basis for Enhanced Affinity and Enzymatic Stability | | Descriptor: | BETA-ALANINE, Presynaptic density protein 95, postsynaptic protein CRIPT peptide | | Authors: | Piserchio, A, Salinas, G.D, Li, T, Marshall, J, Spaller, M.R, Mierke, D.F. | | Deposit date: | 2003-11-12 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Targeting Specific PDZ Domains of PSD-95; Structural Basis for Enhanced Affinity and Enzymatic Stability of a Cyclic Peptide.
Chem.Biol., 11, 2004
|
|
1RGS
 
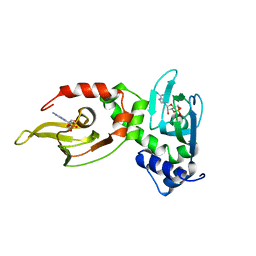 | | REGULATORY SUBUNIT OF CAMP DEPENDENT PROTEIN KINASE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CAMP DEPENDENT PROTEIN KINASE | | Authors: | Su, Y, Dostmann, W.R.G, Herberg, F.W, Durick, K, Xuong, N.-H, Ten Eyck, L, Taylor, S.S, Varughese, K.I. | | Deposit date: | 1995-06-21 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding domains.
Science, 269, 1995
|
|
1RGT
 
 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine, and tetranucleotide AGTC | | Descriptor: | 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*AP*GP*TP*C)-3', ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-12 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
1RGU
 
 | | The crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine, and tetranucleotide AGTG | | Descriptor: | 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*AP*GP*TP*G)-3', SPERMINE, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-13 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
1RGV
 
 | | Crystal Structure of the Ferredoxin from Thauera aromatica | | Descriptor: | IRON/SULFUR CLUSTER, ferredoxin | | Authors: | Unciuleac, M, Boll, M, Warkentin, E, Ermler, U. | | Deposit date: | 2003-11-13 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallization of 4-hydroxybenzoyl-CoA reductase and the structure of its electron donor ferredoxin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1RGW
 
 | | Solution Structure of ZASP's PDZ domain | | Descriptor: | ZASP protein | | Authors: | Au, Y, Atkinson, R.A, Pallavicini, A, Joseph, C, Martin, S.R, Muskett, F.W, Guerrini, R, Faulkner, G, Pastore, A. | | Deposit date: | 2003-11-13 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of ZASP PDZ Domain; Implications for Sarcomere Ultrastructure and Enigma Family Redundancy.
Structure, 12, 2004
|
|
1RGX
 
 | | Crystal Structure of resisitin | | Descriptor: | Resistin | | Authors: | Patel, S.D, Rajala, M.W, Scherer, P.E, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-13 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Disulfide-dependent multimeric assembly of resistin family hormones
Science, 304, 2004
|
|
1RGY
 
 | | Citrobacter freundii GN346 Class C beta-Lactamase Complexed with Transition-State Analog of Cefotaxime | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, beta-lactamase, {[(2E)-2-(2-AMINO-1,3-THIAZOL-4-YL)-2-(METHOXYIMINO)ETHANOYL]AMINO}METHYLPHOSPHONIC ACID | | Authors: | Nukaga, M, Kumar, S, Nukaga, K, Pratt, R.F, Knox, J.R. | | Deposit date: | 2003-11-13 | | Release date: | 2004-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Hydrolysis of third-generation cephalosporins by class C beta-lactamases. Structures of a transition state analog of cefotoxamine in wild-type and extended spectrum enzymes.
J.Biol.Chem., 279, 2004
|
|
1RGZ
 
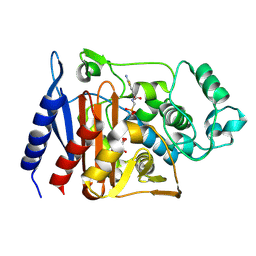 | | Enterobacter cloacae GC1 Class C beta-Lactamase Complexed with Transition-State Analog of Cefotaxime | | Descriptor: | GLYCEROL, class C beta-lactamase, {[(2E)-2-(2-AMINO-1,3-THIAZOL-4-YL)-2-(METHOXYIMINO)ETHANOYL]AMINO}METHYLPHOSPHONIC ACID | | Authors: | Nukaga, M, Kumar, S, Nukaga, K, Pratt, R.F, Knox, J.R. | | Deposit date: | 2003-11-13 | | Release date: | 2004-04-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Hydrolysis of third-generation cephalosporins by class C beta-lactamases. Structures of a transition state analog of cefotoxamine in wild-type and extended spectrum enzymes.
J.Biol.Chem., 279, 2004
|
|
1RH0
 
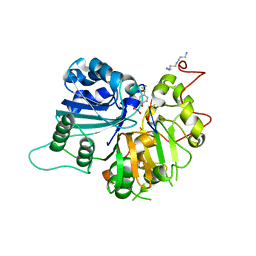 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine and trinucleotide GTT | | Descriptor: | 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*GP*TP*T)-3', SPERMINE, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-13 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
1RH1
 
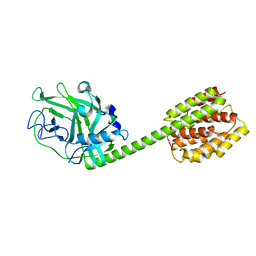 | | crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution | | Descriptor: | Colicin B | | Authors: | Hilsenbeck, J.L, Park, H, Chen, G, Youn, B, Postle, K, Kang, C. | | Deposit date: | 2003-11-13 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution
Mol.Microbiol., 51, 2004
|
|
1RH2
 
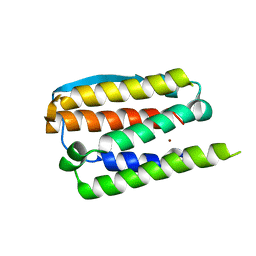 | | RECOMBINANT HUMAN INTERFERON-ALPHA 2B | | Descriptor: | INTERFERON-ALPHA 2B, ZINC ION | | Authors: | Walter, M.R. | | Deposit date: | 1996-11-07 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Zinc mediated dimer of human interferon-alpha 2b revealed by X-ray crystallography.
Structure, 4, 1996
|
|
1RH3
 
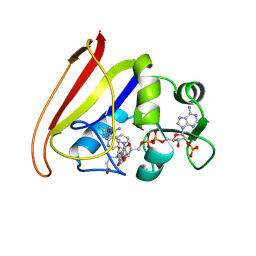 | |
1RH4
 
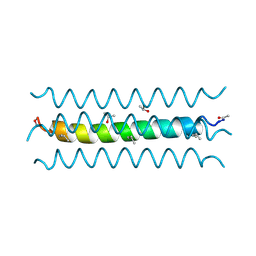 | | RH4 DESIGNED RIGHT-HANDED COILED COIL TETRAMER | | Descriptor: | ISOPROPYL ALCOHOL, RIGHT-HANDED COILED COIL TETRAMER | | Authors: | Harbury, P.B, Plecs, J.J, Tidor, B, Alber, T, Kim, P.S. | | Deposit date: | 1998-10-07 | | Release date: | 1998-12-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution protein design with backbone freedom.
Science, 282, 1998
|
|
1RH5
 
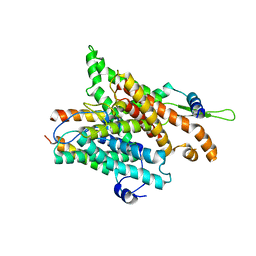 | | The structure of a protein conducting channel | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, SecBeta | | Authors: | van den Berg, B, Clemons Jr, W.M, Collinson, I, Modis, Y, Hartmann, E, Harrison, S.C, Rapoport, T.A. | | Deposit date: | 2003-11-13 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a protein-conducting channel
Nature, 427, 2004
|
|
1RH6
 
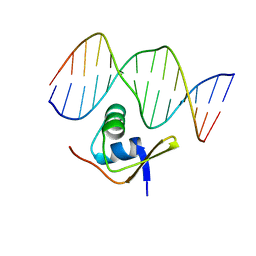 | | Bacteriophage Lambda Excisionase (Xis)-DNA Complex | | Descriptor: | 5'-D(*CP*TP*AP*TP*GP*TP*AP*GP*TP*CP*TP*GP*TP*TP*G)-3', 5'-D(P*CP*AP*AP*CP*AP*GP*AP*CP*TP*AP*CP*AP*TP*AP*G)-3', Excisionase | | Authors: | Sam, M.D, Cascio, D, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2003-11-13 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the excisionase-DNA complex from bacteriophage lambda.
J.Mol.Biol., 338, 2004
|
|
1RH7
 
 | | Crystal Structure of Resistin-like beta | | Descriptor: | HEXAETHYLENE GLYCOL, PLATINUM (II) ION, Resistin-like beta | | Authors: | Patel, S.D, Rajala, M.W, Scherer, P.E, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-13 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Disulfide-dependent multimeric assembly of resistin family hormones
Science, 304, 2004
|
|
