1OWN
 
 | | DATA3:DNA photolyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWO
 
 | | DATA4:photoreduced DNA photolyase / received X-rays dose 1.2 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWP
 
 | | DATA6:photoreduced DNA pholyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWQ
 
 | | Crystal structure of a 40 kDa signalling protein (SPC-40) secreted during involution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, signal processing protein | | Authors: | Kumar, J, Sharma, S, Jasti, J, Bhushan, A, Singh, T.P. | | Deposit date: | 2003-03-29 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a 40 kDa signalling protein (SPC-40) secreted during involution
To be Published
|
|
1OWR
 
 | |
1OWS
 
 | | Crystal structure of a C49 Phospholipase A2 from Indian cobra reveals carbohydrate binding in the hydrophobic channel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Phospholipase A2, ZINC ION | | Authors: | Jabeen, T, Jasti, J, Singh, N, Singh, R.K, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2003-03-30 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a C49 Phospholipase A2 from Indian cobra reveals carbohydrate binding in the hydrophobic channel
To be Published
|
|
1OWT
 
 | | Structure of the Alzheimer's disease amyloid precursor protein copper binding domain | | Descriptor: | Amyloid beta A4 protein | | Authors: | Barnham, K.J, McKinstry, W.J, Multhaup, G, Galatis, D, Morton, C.J, Curtain, C.C, Williamson, N.A, White, A.R, Hinds, M.G, Norton, R.S, Beyreuther, K, Masters, C.L, Parker, M.W, Cappai, R. | | Deposit date: | 2003-03-30 | | Release date: | 2003-05-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the Alzheimer's Disease Amyloid Precursor Protein Copper Binding Domain. A REGULATOR OF NEURONAL COPPER HOMEOSTASIS.
J.Biol.Chem., 278, 2003
|
|
1OWW
 
 | | Solution structure of the first type III module of human fibronectin determined by 1H, 15N NMR spectroscopy | | Descriptor: | Fibronectin first type III module | | Authors: | Gao, M, Craig, D, Lequin, O, Campbell, I.D, Vogel, V, Schulten, K. | | Deposit date: | 2003-03-31 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and functional significance of mechanically unfolded fibronectin type III1 intermediates
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1OWX
 
 | | Solution structure of the C-terminal RRM of human La (La225-334) | | Descriptor: | Lupus La protein | | Authors: | Jacks, A, Babon, J, Kelly, G, Manolaridis, I, Cary, P.D, Curry, S, Conte, M.R. | | Deposit date: | 2003-03-31 | | Release date: | 2003-07-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element
Structure, 11, 2003
|
|
1OWY
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound With 2-Propyl-Aniline | | Descriptor: | 2-PROPYL-ANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-31 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1OWZ
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound With 4-FluoroPhenEthyl Alcohol | | Descriptor: | 4-FLUOROPHENETHYL ALCOHOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-31 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1OX0
 
 | |
1OX1
 
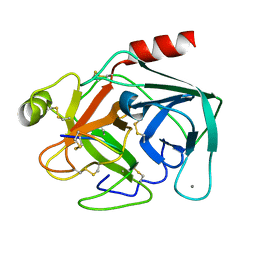 | | crystal structure of the bovine trypsin complex with a synthetic 11 peptide inhibitor | | Descriptor: | 11-mer peptide, CALCIUM ION, Trypsinogen, ... | | Authors: | Wu, G, Huang, Y, Zhu, G, Huang, Q, Tang, Y, Miyake, H, Kusunoki, M. | | Deposit date: | 2003-03-31 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of the bovine trypsin complex with a synthetic 11 peptide inhibitor
To be published
|
|
1OX3
 
 | |
1OX4
 
 | |
1OX5
 
 | |
1OX6
 
 | |
1OX7
 
 | |
1OX8
 
 | | Crystal structure of SspB | | Descriptor: | Stringent starvation protein B | | Authors: | Song, H.K, Eck, M.J. | | Deposit date: | 2003-04-01 | | Release date: | 2003-08-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of degradation signal recognition by SspB, a specificity-enhancing factor for the ClpXP proteolytic machine
Mol.Cell, 12, 2003
|
|
1OX9
 
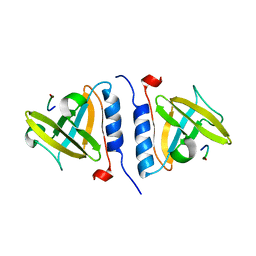 | | Crystal structure of SspB-ssrA complex | | Descriptor: | Stringent starvation protein B, ssrA | | Authors: | Song, H.K, Eck, M.J. | | Deposit date: | 2003-04-01 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of degradation signal recognition by SspB, a specificity-enhancing factor for the ClpXP proteolytic machine
Mol.Cell, 12, 2003
|
|
1OXA
 
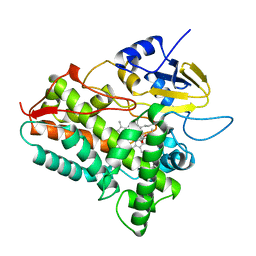 | | CYTOCHROME P450 (DONOR:O2 OXIDOREDUCTASE) | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, CYTOCHROME P450 ERYF, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cupp-Vickery, J.R, Poulos, T.L. | | Deposit date: | 1995-07-14 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of cytochrome P450eryF involved in erythromycin biosynthesis.
Nat.Struct.Biol., 2, 1995
|
|
1OXB
 
 | |
1OXC
 
 | | LecB (PA-LII) in complex with FUCOSE | | Descriptor: | CALCIUM ION, SULFATE ION, alpha-L-fucopyranose, ... | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-04-02 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
1OXD
 
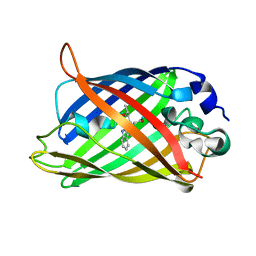 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
1OXE
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
