1JVN
 
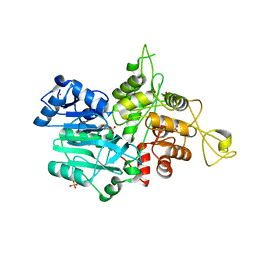 | | CRYSTAL STRUCTURE OF IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE: A TUNNEL THROUGH A (BETA/ALPHA)8 BARREL JOINS TWO ACTIVE SITES | | Descriptor: | BIFUNCTIONAL HISTIDINE BIOSYNTHESIS PROTEIN HISHF, NICKEL (II) ION, PYROPHOSPHATE 2-, ... | | Authors: | Chaudhuri, B.N, Smith, J.L, Davisson, V.J, Myers, R.S, Lange, S.C, Chittur, S.V. | | Deposit date: | 2001-08-30 | | Release date: | 2001-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of imidazole glycerol phosphate synthase: a tunnel through a (beta/alpha)8 barrel joins two active sites.
Structure, 9, 2001
|
|
1JVO
 
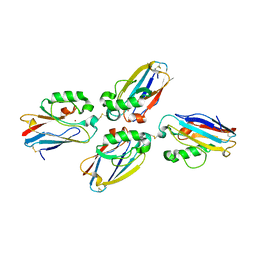 | | Azurin dimer, crosslinked via disulfide bridge | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | van Amsterdam, I.M.C, Ubbink, M, Einsle, O, Messerschmidt, A, Merli, A, Cavazzini, D, Rossi, G.L, Canters, G.W. | | Deposit date: | 2001-08-30 | | Release date: | 2002-01-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Dramatic modulation of electron transfer in protein complexes by crosslinking
Nat.Struct.Biol., 9, 2002
|
|
1JVP
 
 | |
1JVQ
 
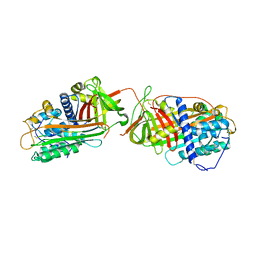 | | Crystal structure at 2.6A of the ternary complex between antithrombin, a P14-P8 reactive loop peptide, and an exogenous tetrapeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, ... | | Authors: | Zhou, A, Huntington, J.A, Lomas, D.A, Carrell, R.W, Stein, P.E. | | Deposit date: | 2001-08-31 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | How small peptides block and reverse serpin polymerisation
J.Mol.Biol., 342, 2004
|
|
1JVR
 
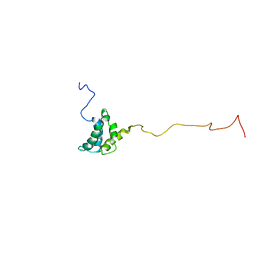 | | STRUCTURE OF THE HTLV-II MATRIX PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | HUMAN T-CELL LEUKEMIA VIRUS TYPE II MATRIX PROTEIN | | Authors: | Christensen, A.M, Massiah, M.A, Turner, B.G, Sundquist, W.I, Summers, M.F. | | Deposit date: | 1996-10-03 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the HTLV-II matrix protein and comparative analysis of matrix proteins from the different classes of pathogenic human retroviruses.
J.Mol.Biol., 264, 1996
|
|
1JVS
 
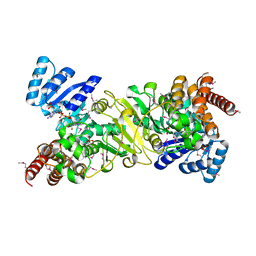 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase; a target enzyme for antimalarial drugs | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Yajima, S, Nonaka, T, Kuzuyama, T, Seto, H, Ohsawa, K. | | Deposit date: | 2001-08-31 | | Release date: | 2002-10-09 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase complexed with cofactors: implications of a flexible loop movement upon substrate binding.
J.Biochem., 131, 2002
|
|
1JVT
 
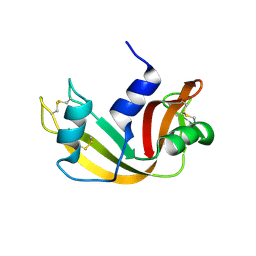 | |
1JVU
 
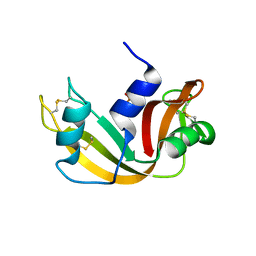 | | CRYSTAL STRUCTURE OF RIBONUCLEASE A (COMPLEXED FORM) | | Descriptor: | CYTIDINE-2'-MONOPHOSPHATE, RIBONUCLEASE A | | Authors: | Vitagliano, L, Merlino, A, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-08-31 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Reversible Substrate-Induced Domain Motions in Ribonuclease A
Proteins, 46, 2002
|
|
1JVV
 
 | |
1JVW
 
 | | TRYPANOSOMA CRUZI MACROPHAGE INFECTIVITY POTENTIATOR (TCMIP) | | Descriptor: | MACROPHAGE INFECTIVITY POTENTIATOR | | Authors: | Pereira, P.J.B, Vega, M.C, Gonzalez-Rey, E, Fernandez-Carazo, R, Macedo-Ribeiro, S, Gomis-Rueth, F.X, Gonzalez, A, Coll, M. | | Deposit date: | 2001-08-31 | | Release date: | 2002-06-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Trypanosoma cruzi macrophage infectivity potentiator has a rotamase core and a highly exposed alpha-helix.
EMBO Rep., 3, 2002
|
|
1JVX
 
 | | Maltodextrin-binding protein variant D207C/A301GS/P316C cross-linked in crystal | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltodextrin-binding protein | | Authors: | Srinivasan, U, Iyer, G.H, Przybycien, T.A, Samsonoff, W.A, Bell, J.A. | | Deposit date: | 2001-08-31 | | Release date: | 2001-09-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystine: Fibrous Biomolecular Material from Protein Crystals Cross-linked in a Specific Geometry
Protein Eng., 15, 2002
|
|
1JVY
 
 | | Maltodextrin-binding protein variant D207C/A301GS/P316C with beta-mercaptoethanol mixed disulfides | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltodextrin-binding protein | | Authors: | Srinivasan, U, Iyer, G.H, Przybycien, T.A, Samsonoff, W.A, Bell, J.A. | | Deposit date: | 2001-08-31 | | Release date: | 2001-09-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystine: Fibrous Biomolecular Material from Protein Crystals Cross-linked in a Specific Geometry
Protein Eng., 15, 2002
|
|
1JVZ
 
 | | Structure of cephalosporin acylase in complex with glutaryl-7-aminocephalosporanic acid | | Descriptor: | 7BETA-(4CARBOXYBUTANAMIDO) CEPHALOSPORANIC ACID, cephalosporin acylase alpha chain, cephalosporin acylase beta chain | | Authors: | Kim, Y, Hol, W.G.J. | | Deposit date: | 2001-09-01 | | Release date: | 2002-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of cephalosporin acylase in complex with glutaryl-7-aminocephalosporanic acid and glutarate: insight into the basis of its substrate specificity
CHEM.BIOL., 8, 2001
|
|
1JW0
 
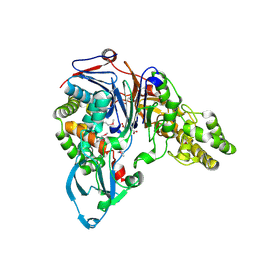 | | Structure of cephalosporin acylase in complex with glutarate | | Descriptor: | GLUTARIC ACID, cephalosporin acylase alpha chain, cephalosporin acylase beta chain | | Authors: | Kim, Y, Hol, W.G.J. | | Deposit date: | 2001-09-01 | | Release date: | 2002-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of cephalosporin acylase in complex with glutaryl-7-aminocephalosporanic acid and glutarate: insight into the basis of its substrate specificity
CHEM.BIOL., 8, 2001
|
|
1JW1
 
 | |
1JW2
 
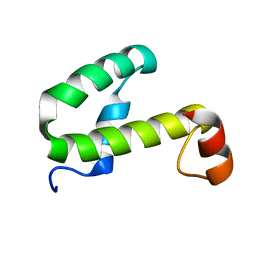 | | SOLUTION STRUCTURE OF HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha FROM ESCHERICHIA COLI. Ontario Centre for Structural Proteomics target EC0308_1_72; Northeast Structural Genomics Target ET88 | | Descriptor: | HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha | | Authors: | Chang, X, Yee, A, Savchenko, A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JW3
 
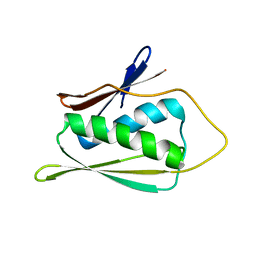 | | Solution Structure of Methanobacterium Thermoautotrophicum Protein 1598. Ontario Centre for Structural Proteomics target MTH1598_1_140; Northeast Structural Genomics Target TT6 | | Descriptor: | Conserved Hypothetical Protein MTH1598 | | Authors: | Chang, X, Connelly, G, Yee, A, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JW4
 
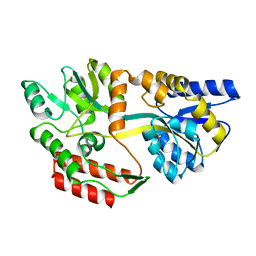 | | Structure of ligand-free maltodextrin-binding protein | | Descriptor: | maltodextrin-binding protein | | Authors: | Duan, X, Quiocho, F.A. | | Deposit date: | 2001-09-02 | | Release date: | 2002-01-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for a dominant role of nonpolar interactions in the binding of a transport/chemosensory receptor to its highly polar ligands.
Biochemistry, 41, 2002
|
|
1JW5
 
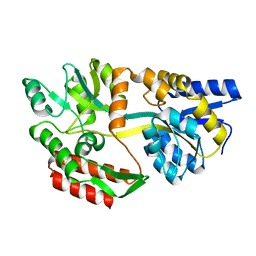 | |
1JW6
 
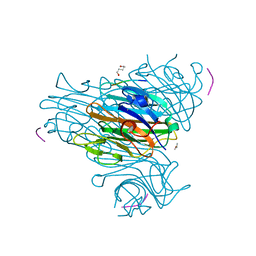 | | Crystal Structure of the Complex of Concanavalin A and Hexapeptide | | Descriptor: | CALCIUM ION, Concanavalin A, ISOPROPYL ALCOHOL, ... | | Authors: | Zhang, Z, Qian, M, Huang, Q, Jia, Y, Tang, Y. | | Deposit date: | 2001-09-02 | | Release date: | 2001-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the complex of concanavalin A and hexapeptide.
J.Protein Chem., 20, 2001
|
|
1JW8
 
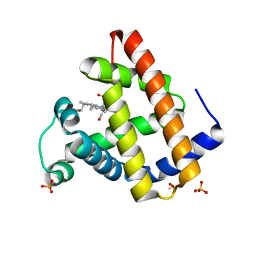 | | 1.3 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF P6 FORM OF MYOGLOBIN | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Phillips Jr, G.N. | | Deposit date: | 2001-09-03 | | Release date: | 2001-10-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sampling of the native conformational ensemble of myoglobin via structures in different crystalline environments.
Proteins, 70, 2008
|
|
1JW9
 
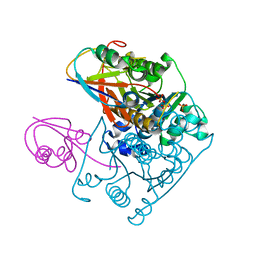 | | Structure of the Native MoeB-MoaD Protein Complex | | Descriptor: | MOLYBDOPTERIN BIOSYNTHESIS MOEB PROTEIN, MOLYBDOPTERIN [MPT] CONVERTING FACTOR, SUBUNIT 1, ... | | Authors: | Lake, M.W, Wuebbens, M.M, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2001-09-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex.
Nature, 414, 2001
|
|
1JWA
 
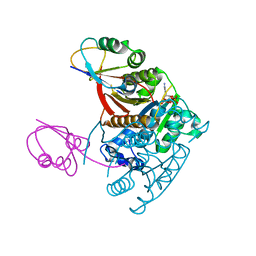 | | Structure of the ATP-bound MoeB-MoaD Protein Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MOLYBDOPTERIN BIOSYNTHESIS MOEB PROTEIN, MOLYBDOPTERIN [MPT] CONVERTING FACTOR, ... | | Authors: | Lake, M.W, Wuebbens, M.M, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2001-09-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex.
Nature, 414, 2001
|
|
1JWB
 
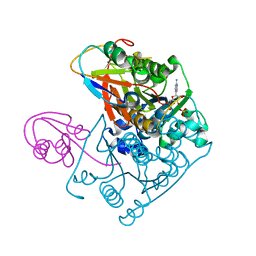 | | Structure of the Covalent Acyl-Adenylate Form of the MoeB-MoaD Protein Complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, MOLYBDOPTERIN BIOSYNTHESIS MOEB PROTEIN, MOLYBDOPTERIN [MPT] CONVERTING FACTOR, ... | | Authors: | Lake, M.W, Wuebbens, M.M, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2001-09-03 | | Release date: | 2001-11-21 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex.
Nature, 414, 2001
|
|
1JWC
 
 | |
