1GP1
 
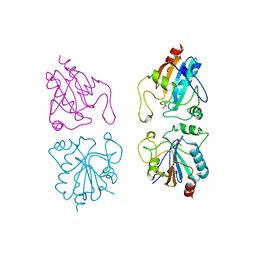 | |
1GP2
 
 | |
1GP3
 
 | | Human IGF2R domain 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Brown, J, Esnouf, R.M, Jones, M.A, Linnell, J, Harlos, K, Hassan, A.B, Jones, E.Y. | | Deposit date: | 2001-10-29 | | Release date: | 2002-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a Functional Igf2R Fragment Determined from the Anomalous Scattering of Sulfur
Embo J., 21, 2002
|
|
1GP4
 
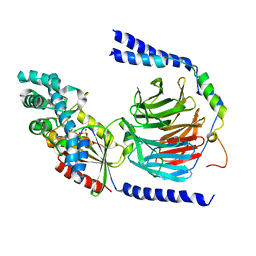
 | | Anthocyanidin synthase from Arabidopsis thaliana (selenomethionine substituted) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-OXOGLUTARIC ACID, ANTHOCYANIDIN SYNTHASE | | Authors: | Wilmouth, R.C, Turnbull, J.J, Welford, R.W.D, Clifton, I.J, Prescott, A.G, Schofield, C.J. | | Deposit date: | 2001-10-30 | | Release date: | 2002-02-21 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Mechanism of Anthocyanidin Synthase from Arabidopsis Thaliana.
Structure, 10, 2002
|
|
1GP5
 
 | | Anthocyanidin synthase from Arabidopsis thaliana complexed with trans-dihydroquercetin | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, (2S,3S)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Wilmouth, R.C, Turnbull, J.J, Welford, R.W.D, Clifton, I.J, Prescott, A.G, Schofield, C.J. | | Deposit date: | 2001-10-30 | | Release date: | 2002-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Mechanism of Anthocyanidin Synthase from Arabidopsis Thaliana.
Structure, 10, 2002
|
|
1GP6
 
 | | Anthocyanidin synthase from Arabidopsis thaliana complexed with trans-dihydroquercetin (with 30 min exposure to O2) | | Descriptor: | (2S,3S)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,5,7,3',4'-PENTAHYDROXYFLAVONE, ... | | Authors: | Wilmouth, R.C, Turnbull, J.J, Welford, R.W.D, Clifton, I.J, Prescott, A.G, Schofield, C.J. | | Deposit date: | 2001-10-30 | | Release date: | 2002-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Mechanism of Anthocyanidin Synthase from Arabidopsis Thaliana.
Structure, 10, 2002
|
|
1GP7
 
 | |
1GP8
 
 | | NMR SOLUTION STRUCTURE OF THE COAT PROTEIN-BINDING DOMAIN OF BACTERIOPHAGE P22 SCAFFOLDING PROTEIN | | Descriptor: | PROTEIN (SCAFFOLDING PROTEIN) | | Authors: | Sun, Y, Parker, M.H, Weigele, P, Casjens, S, Prevelige Jr, P.E, Krishna, N.R. | | Deposit date: | 1999-05-11 | | Release date: | 1999-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus.
J.Mol.Biol., 297, 2000
|
|
1GP9
 
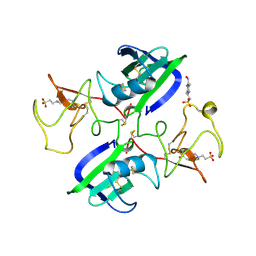 | | A New Crystal Form of the Nk1 Splice Variant of Hgf/Sf Demonstrates Extensive Hinge Movement and Suggests that the Nk1 Dimer Originates by Domain Swapping | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEPATOCYTE GROWTH FACTOR | | Authors: | Watanabe, K, Chirgadze, D.Y, Lietha, D, Gherardi, E, Blundell, T.L. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A New Crystal Form of the Nk1 Splice Variant of Hgf/Sf Demonstrates Extensive Hinge Movement and Suggests that the Nk1 Dimer Originates by Domain Swapping
J.Mol.Biol., 319, 2002
|
|
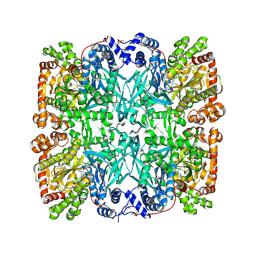
1GPA
 
 | |
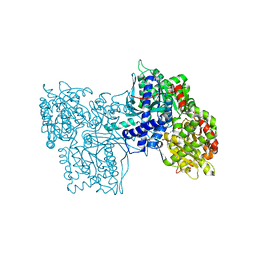
1GPB
 
 | |
1GPC
 
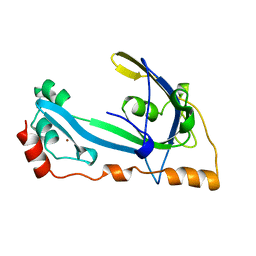 | | CORE GP32, DNA-BINDING PROTEIN | | Descriptor: | PROTEIN (CORE GP32), ZINC ION | | Authors: | Shamoo, Y, Friedman, A.M, Parsons, M.R, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1995-06-01 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA.
Nature, 376, 1995
|
|
1GPD
 
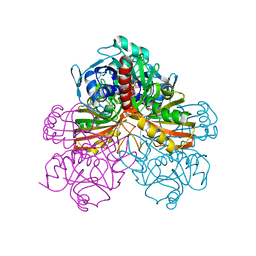 | | STUDIES OF ASYMMETRY IN THE THREE-DIMENSIONAL STRUCTURE OF LOBSTER D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Moras, D, Olsen, K.W, Sabesan, M.N, Buehner, M, Ford, G.C, Rossmann, M.G. | | Deposit date: | 1975-07-01 | | Release date: | 1977-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Studies of asymmetry in the three-dimensional structure of lobster D-glyceraldehyde-3-phosphate dehydrogenase.
J.Biol.Chem., 250, 1975
|
|
1GPE
 
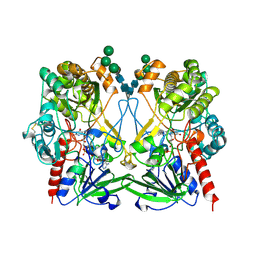 | | GLUCOSE OXIDASE FROM PENICILLIUM AMAGASAKIENSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hendle, J, Kalisz, H.M, Hecht, H.J. | | Deposit date: | 1999-03-24 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 and 1.9 A resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1GPF
 
 | |
1GPH
 
 | |
1GPI
 
 | |
1GPJ
 
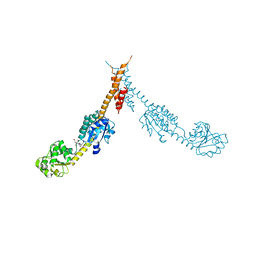 | | Glutamyl-tRNA Reductase from Methanopyrus kandleri | | Descriptor: | (2R,3R,4S,5S)-4-AMINO-2-[6-(DIMETHYLAMINO)-9H-PURIN-9-YL]-5-(HYDROXYMETHYL)TETRAHYDRO-3-FURANOL, CITRIC ACID, GLUTAMIC ACID, ... | | Authors: | Moser, J, Schubert, W.-D, Beier, V, Bringemeier, I, Jahn, D, Heinz, D.W. | | Deposit date: | 2001-11-05 | | Release date: | 2002-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis.
EMBO J., 20, 2001
|
|
1GPK
 
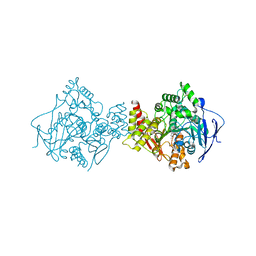 | | Structure of Acetylcholinesterase Complex with (+)-Huperzine A at 2.1A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Dvir, H, Harel, M, Chetrit, M, Silman, I, Sussman, J.L. | | Deposit date: | 2001-11-05 | | Release date: | 2002-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Structures of Torpedo Californica Acetylcholinesterase Complexed with (+)-Huperzine a and (-)-Huperzine B: Structural Evidence for an Active Site Rearrangement
Biochemistry, 41, 2002
|
|
1GPL
 
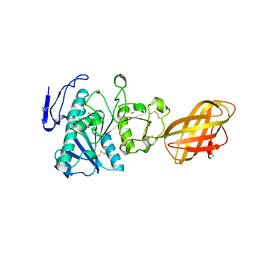 | | RP2 LIPASE | | Descriptor: | CALCIUM ION, RP2 LIPASE | | Authors: | Withers-Martinez, C, Cambillau, C. | | Deposit date: | 1996-07-13 | | Release date: | 1997-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A pancreatic lipase with a phospholipase A1 activity: crystal structure of a chimeric pancreatic lipase-related protein 2 from guinea pig.
Structure, 4, 1996
|
|
1GPM
 
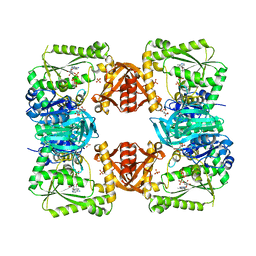 | | ESCHERICHIA COLI GMP SYNTHETASE COMPLEXED WITH AMP AND PYROPHOSPHATE | | Descriptor: | ADENOSINE MONOPHOSPHATE, CITRIC ACID, GMP SYNTHETASE, ... | | Authors: | Tesmer, J.J.G. | | Deposit date: | 1995-04-04 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of GMP synthetase reveals a novel catalytic triad and is a structural paradigm for two enzyme families.
Nat.Struct.Biol., 3, 1996
|
|
1GPN
 
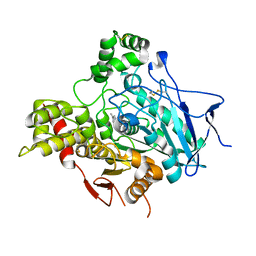 | | STRUCTURE OF ACETYLCHOLINESTERASE COMPLEXED WITH HUPERZINE B AT 2.35A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, HUPERZINE B | | Authors: | Dvir, H, Harel, M, Jiang, H.L, Silman, I, Sussman, J.L. | | Deposit date: | 2001-11-07 | | Release date: | 2002-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-Ray Structures of Torpedo Californica Acetylcholinesterase Complexed with (+)-Huperzine a and (-)-Huperzine B: Structural Evidence for an Active Site Rearrangement
Biochemistry, 41, 2002
|
|
1GPO
 
 | |
1GPP
 
 | |
1GPQ
 
 | | Structure of ivy complexed with its target, HEWL | | Descriptor: | INHIBITOR OF VERTEBRATE LYSOZYME, LYSOZYME C | | Authors: | Abergel, C, Monchois, V, Claverie, J.-M. | | Deposit date: | 2001-11-08 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Evolution of the Ivy Protein Family, Unexpected Lysozyme Inhibitors in Gram-Negative Bacteria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
