1AVW
 
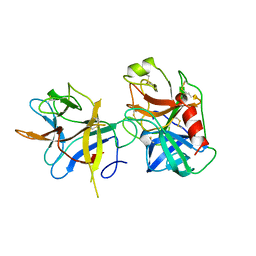 | | COMPLEX PORCINE PANCREATIC TRYPSIN/SOYBEAN TRYPSIN INHIBITOR, ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-21 | | Release date: | 1998-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kunitz-type soybean trypsin inhibitor revisited: refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator.
J.Mol.Biol., 275, 1998
|
|
1AVX
 
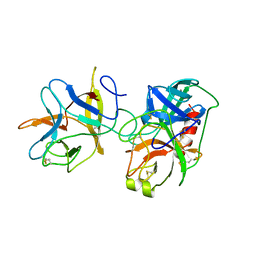 | | COMPLEX PORCINE PANCREATIC TRYPSIN/SOYBEAN TRYPSIN INHIBITOR, TETRAGONAL CRYSTAL FORM | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-21 | | Release date: | 1998-10-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kunitz-type soybean trypsin inhibitor revisited: refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator.
J.Mol.Biol., 275, 1998
|
|
1AVY
 
 | | FIBRITIN DELETION MUTANT M (BACTERIOPHAGE T4) | | Descriptor: | FIBRITIN | | Authors: | Strelkov, S.V, Tao, Y, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 1997-09-22 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of bacteriophage T4 fibritin M: a troublesome packing arrangement.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AVZ
 
 | |
1AW0
 
 | |
1AW1
 
 | |
1AW2
 
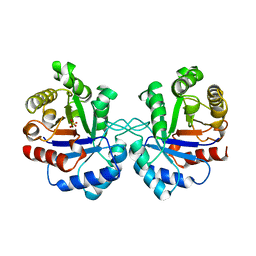 | | TRIOSEPHOSPHATE ISOMERASE OF VIBRIO MARINUS | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Maes, D, Zeelen, J.P, Wierenga, R.K. | | Deposit date: | 1997-10-09 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Triose-phosphate isomerase (TIM) of the psychrophilic bacterium Vibrio marinus. Kinetic and structural properties.
J.Biol.Chem., 273, 1998
|
|
1AW3
 
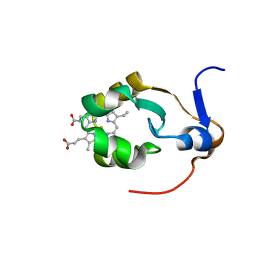 | | THE SOLUTION NMR STRUCTURE OF OXIDIZED RAT MICROSOMAL CYTOCHROME B5, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Felli, I.C. | | Deposit date: | 1997-10-09 | | Release date: | 1998-02-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of oxidized rat microsomal cytochrome b5.
Biochemistry, 37, 1998
|
|
1AW4
 
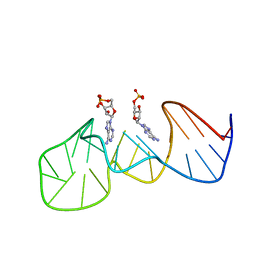 | |
1AW5
 
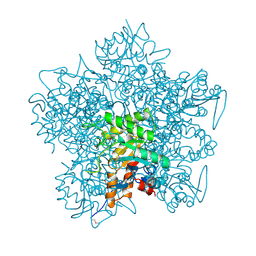 | |
1AW6
 
 | | GAL4 (CD), NMR, 24 STRUCTURES | | Descriptor: | CADMIUM ION, GAL4 (CD) | | Authors: | Baleja, J.D, Wagner, G. | | Deposit date: | 1997-10-10 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the DNA-binding domain of GAL4 and use of 3J(113Cd,1H) in structure determination.
J.Biomol.NMR, 10, 1997
|
|
1AW7
 
 | | Q136A MUTANT OF TOXIC SHOCK SYNDROME TOXIN-1 FROM S. AUREUS | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Earhart, C.A, Mitchell, D.T, Murray, D.L, Pinheiro, D.M, Matsumura, M, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1997-10-11 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity.
Biochemistry, 37, 1998
|
|
1AW8
 
 | | PYRUVOYL DEPENDENT ASPARTATE DECARBOXYLASE | | Descriptor: | L-ASPARTATE-ALPHA-DECARBOXYLASE | | Authors: | Albert, A, Dhanaraj, V, Genschel, U, Khan, G, Ramjee, M.K, Pulido, R, Sybanda, B.L, von Delf, F, Witty, M, Blundell, T.L, Smith, A.G, Abell, C. | | Deposit date: | 1997-10-12 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of aspartate decarboxylase at 2.2 A resolution provides evidence for an ester in protein self-processing.
Nat.Struct.Biol., 5, 1998
|
|
1AW9
 
 | | STRUCTURE OF GLUTATHIONE S-TRANSFERASE III IN APO FORM | | Descriptor: | CADMIUM ION, GLUTATHIONE S-TRANSFERASE III | | Authors: | Neuefeind, T, Huber, R, Reinemer, P, Knaeblein, J. | | Deposit date: | 1997-10-13 | | Release date: | 1998-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cloning, sequencing, crystallization and X-ray structure of glutathione S-transferase-III from Zea mays var. mutin: a leading enzyme in detoxification of maize herbicides.
J.Mol.Biol., 274, 1997
|
|
1AWB
 
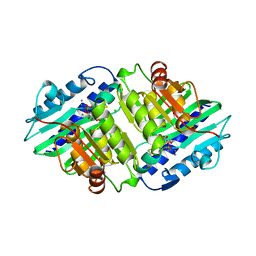 | | HUMAN MYO-INOSITOL MONOPHOSPHATASE IN COMPLEX WITH D-INOSITOL-1-PHOSPHATE AND CALCIUM | | Descriptor: | CALCIUM ION, CHLORIDE ION, D-MYO-INOSITOL-1-PHOSPHATE, ... | | Authors: | Rondeau, J.-M, Pelton, P.D, Vincendon, P, Ganzhorn, A.J. | | Deposit date: | 1997-10-01 | | Release date: | 1998-01-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an Enzyme-Substrate Complex and the Catalytic Mechanism of Human Brain Myo-Inositol Monophosphatase
Protein Eng., 10, 1997
|
|
1AWC
 
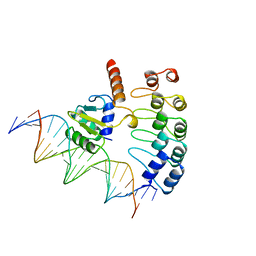 | | MOUSE GABP ALPHA/BETA DOMAIN BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*(BRU)P*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*AP*(CBR)P*AP*CP*(CBR)P*GP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*CP*GP*GP*(BRU)P*GP*(BRU)P*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP*AP*T)-3'), PROTEIN (GA BINDING PROTEIN ALPHA), ... | | Authors: | Batchelor, A.H, Wolberger, C. | | Deposit date: | 1997-10-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of GABPalpha/beta: an ETS domain- ankyrin repeat heterodimer bound to DNA.
Science, 279, 1998
|
|
1AWD
 
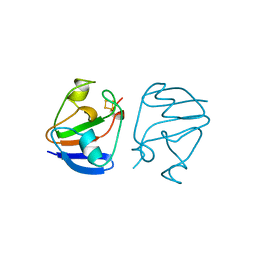 | | FERREDOXIN [2FE-2S] OXIDIZED FORM FROM CHLORELLA FUSCA | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Sheldrick, G.M. | | Deposit date: | 1997-10-01 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure determination at 1.4 A resolution of ferredoxin from the green alga Chlorella fusca
Structure Fold.Des., 7, 1999
|
|
1AWE
 
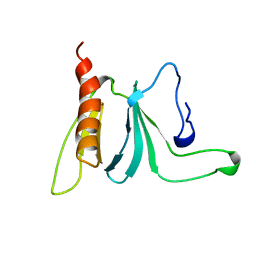 | | HUMAN SOS1 PLECKSTRIN HOMOLOGY (PH) DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | SOS1 | | Authors: | Zheng, J, Cowburn, D. | | Deposit date: | 1997-10-01 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the pleckstrin homology domain of human SOS1. A possible structural role for the sequential association of diffuse B cell lymphoma and pleckstrin homology domains.
J.Biol.Chem., 272, 1997
|
|
1AWF
 
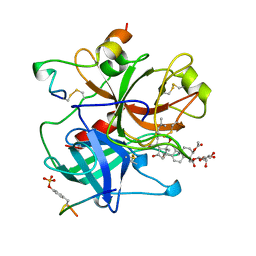 | | NOVEL COVALENT THROMBIN INHIBITOR FROM PLANT EXTRACT | | Descriptor: | ALPHA THROMBIN, HIRUGEN, R3-ACETOXY-17-(1-FORMYL-5-METHYL-3-OXO-HEX-4-ENYL)-12,16-DIHYDROXY-14-HYDROXYMETHYL-4,10,13-TRIMETHYL-2,3,4,5,6,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYCLOPENTA[A]PHENANTHRENE-4-CARBOXYLIC ACID IDOPYRANOSYL ESTER | | Authors: | Jhoti, H, Cleasby, A, Wonacott, A. | | Deposit date: | 1997-10-02 | | Release date: | 1998-10-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel natural product 5,5-trans-lactone inhibitors of human alpha-thrombin: mechanism of action and structural studies.
Biochemistry, 37, 1998
|
|
1AWH
 
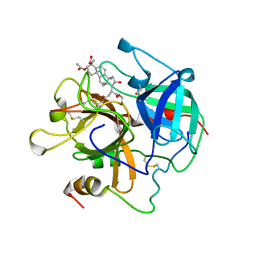 | | NOVEL COVALENT THROMBIN INHIBITOR FROM PLANT EXTRACT | | Descriptor: | 3-ACETOXY-17-(1-FORMYL-5-METHYL-3-OXO-HEX-4-ENYL)-16-HYDROXY-4,10,13,14-TETRAMETHYL-2,3,4,5,6,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYCLOPENTA[A]PHENANTHRENE-4-CARBOXYLIC ACID, ALPHA THROMBIN | | Authors: | Jhoti, H, Cleasby, A, Wonacott, A. | | Deposit date: | 1997-10-02 | | Release date: | 1998-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel natural product 5,5-trans-lactone inhibitors of human alpha-thrombin: mechanism of action and structural studies.
Biochemistry, 37, 1998
|
|
1AWI
 
 | |
1AWJ
 
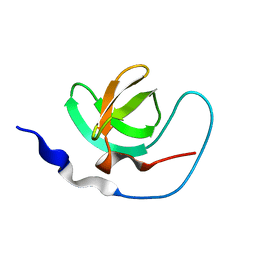 | | INTRAMOLECULAR ITK-PROLINE COMPLEX, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ITK | | Authors: | Andreotti, A.H, Bunnell, S.C, Feng, S, Berg, L.J, Schreiber, S.L. | | Deposit date: | 1997-10-02 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Regulatory intramolecular association in a tyrosine kinase of the Tec family.
Nature, 385, 1997
|
|
1AWO
 
 | |
1AWP
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, X, Zhang, X. | | Deposit date: | 1997-10-03 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The reduction potential of cytochrome b5 is modulated by its exposed heme edge.
Biochemistry, 37, 1998
|
|
1AWQ
 
 | | CYPA COMPLEXED WITH HAGPIA (PSEUDO-SYMMETRIC MONOMER) | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-04 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
