1KCO
 
 | | Structure of e131 Zeta Peptide, a Potent Antagonist of the High-Affinity IgE Receptor | | Descriptor: | e131 Zeta Peptide | | Authors: | Nakamura, G.R, Reynolds, M.E, Chen, Y.M, Starovasnik, M.A, Lowman, H.B. | | Deposit date: | 2001-11-09 | | Release date: | 2002-03-06 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Stable "zeta" peptides that act as potent antagonists of the high-affinity IgE receptor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2BL0
 
 | | Physarum polycephalum myosin II regulatory domain | | Descriptor: | CALCIUM ION, MAJOR PLASMODIAL MYOSIN HEAVY CHAIN, MYOSIN REGULATORY LIGHT CHAIN | | Authors: | Debreczeni, J.E, Farkas, L, Harmat, V, Nyitray, L. | | Deposit date: | 2005-02-23 | | Release date: | 2005-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Evidence for Non-Canonical Binding of Ca2+ to a Canonical EF-Hand of a Conventional Myosin.
J.Biol.Chem., 280, 2005
|
|
1SYI
 
 | | X-RAY STRUCTURE OF THE Y702F MUTANT OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 2.1 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | Authors: | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-04-01 | | Release date: | 2005-03-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
4CCL
 
 | | X-Ray structure of E. coli ycfD | | Descriptor: | 50S RIBOSOMAL PROTEIN L16 ARGININE HYDROXYLASE, MANGANESE (II) ION, SULFATE ION | | Authors: | McDonough, M.A, Ho, C.H, Kershaw, N.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
2R8A
 
 | | Crystal structure of the long-chain fatty acid transporter FadL mutant delta N8 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Long-chain fatty acid transport protein | | Authors: | Hearn, E.M, Patel, D.R, Lepore, B.W, Indic, M, van den Berg, B. | | Deposit date: | 2007-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | From the Cover: Ligand-gated diffusion across the bacterial outer membrane.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8U25
 
 | |
4KKI
 
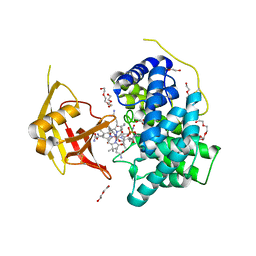 | | Crystal Structure of Haptocorrin in Complex with CNCbl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANOCOBALAMIN, ... | | Authors: | Furger, E, Frei, D.C, Schibli, R, Fischer, E, Prota, A.E. | | Deposit date: | 2013-05-06 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for universal corrinoid recognition by the cobalamin transport protein haptocorrin.
J.Biol.Chem., 288, 2013
|
|
6I8T
 
 | |
7RW9
 
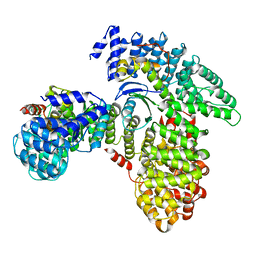 | | AP2 bound to heparin in the bowl conformation | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Baker, R.W, Hollopeter, G, Partlow, E.A. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of an endocytic checkpoint that primes the AP2 clathrin adaptor for cargo internalization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8U4Y
 
 | |
7RW8
 
 | | AP2 bound to heparin in the closed conformation | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Baker, R.W, Hollopeter, G, Partlow, E.A. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of an endocytic checkpoint that primes the AP2 clathrin adaptor for cargo internalization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6I8M
 
 | |
2R89
 
 | | Crystal structure of the long-chain fatty acid transporter FadL mutant delta N3 | | Descriptor: | Long-chain fatty acid transport protein | | Authors: | Hearn, E.M, Patel, D.R, Lepore, B.W, Indic, M, van den Berg, B. | | Deposit date: | 2007-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | From the Cover: Ligand-gated diffusion across the bacterial outer membrane.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6I5M
 
 | | Gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus in complex with GDP and formate ion | | Descriptor: | FORMIC ACID, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kravchenko, O, Nikonov, O, Gabdulkhakov, A, Stolboushkina, E, Arkhipova, V, Garber, M, Nikonov, S. | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The third structural switch in the archaeal translation initiation factor 2 (aIF2) molecule and its possible role in the initiation of GTP hydrolysis and the removal of aIF2 from the ribosome.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4KG2
 
 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with Cefotaxime | | Descriptor: | Beta-lactamase, CEFOTAXIME, C3' cleaved, ... | | Authors: | Docter, B.E, Baggett, V.L, Powers, R.A, Wallar, B.J. | | Deposit date: | 2013-04-28 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Complexed structures of AmpC beta-lactamase
To be Published
|
|
4KG6
 
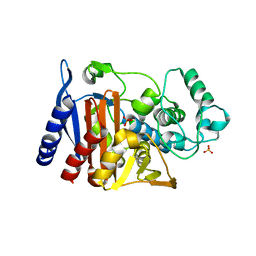 | |
3D00
 
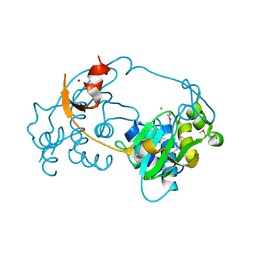 | |
5A7U
 
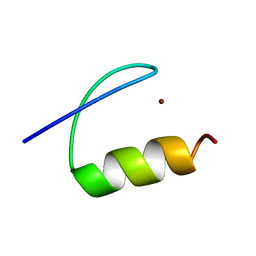 | | Single-particle cryo-EM of co-translational folded adr1 domain inside the E. coli ribosome exit tunnel. | | Descriptor: | REGULATORY PROTEIN ADR1, ZINC ION | | Authors: | Nilsson, O.B, Hedman, R, Marino, J, Wickles, S, Bischoff, L, Johansson, M, Muller-Lucks, A, Trovato, F, Puglisi, J.D, O'Brien, E, Beckmann, R, von Heijne, G. | | Deposit date: | 2015-07-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cotranslational Protein Folding Inside the Ribosome Exit Tunnel.
Cell Rep., 12, 2015
|
|
5K9T
 
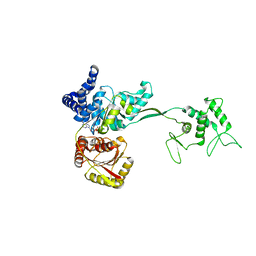 | |
5AGW
 
 | | Bcl-2 alpha beta-1 complex | | Descriptor: | APOPTOSIS REGULATOR BCL-2, BCL-2-LIKE PROTEIN 1, BCL-2-LIKE PROTEIN 11 | | Authors: | Smith, B.J, F Lee, E, Checco, J.W, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-09 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Alpha Beta Peptide Foldamers Targeting Intracellular Protein-Protein Interactions with Activity on Living Cells
J.Am.Chem.Soc., 137, 2015
|
|
5A4K
 
 | | Crystal structure of the R139W variant of human NAD(P)H:quinone oxidoreductase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Strandback, E, Gudipati, V, Uhl, M.K, Rantase, D.M, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2015-06-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Catalytic competence, structure and stability of the cancer-associated R139W variant of the human NAD(P)H:quinone oxidoreductase 1 (NQO1).
FEBS J., 284, 2017
|
|
7BNX
 
 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-22 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1K7H
 
 | | CRYSTAL STRUCTURE OF SHRIMP ALKALINE PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALKALINE PHOSPHATASE, MALEIC ACID, ... | | Authors: | De Backer, M.E, Mc Sweeney, S, Rasmussen, H.B, Riise, B.W, Lindley, P, Hough, E. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The 1.9 A Crystal Structure of Heat-Labile Shrimp Alkaline Phosphatase
J.Mol.Biol., 318, 2002
|
|
7BB5
 
 | |
9FK0
 
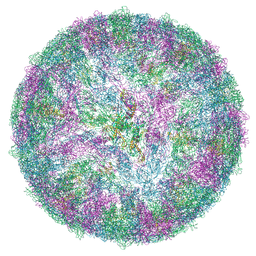 | | LGTV with TBEV prME | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, ... | | Authors: | Bisikalo, K, Rosendal, E. | | Deposit date: | 2024-06-01 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The influence of the pre-membrane and envelope proteins on structure, pathogenicity and tropism of tick-borne encephalitis virus
Biorxiv, 2024
|
|
