4NVJ
 
 | |
3BNE
 
 | | Lipoxygenase-1 (Soybean) I553A Mutant | | Descriptor: | FE (III) ION, Seed lipoxygenase-1 | | Authors: | Tomchick, D.R. | | Deposit date: | 2007-12-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enzyme structure and dynamics affect hydrogen tunneling: the impact of a remote side chain (I553) in soybean lipoxygenase-1.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4O0U
 
 | |
3BXK
 
 | | Crystal structure of the P/Q-type calcium channel (CaV2.1) IQ domain and CA2+calmodulin complex | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Mori, M.X, Vander Kooi, C.W, Leahy, D.J, Yue, D.T. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the P/Q-type calcium channel (CaV2.1) IQ domain and CA2+calmodulin complex
To be Published
|
|
4NC4
 
 | | Crystal structure of photoreceptor AtUVR8 mutant W285F and light-induced structural changes at 120K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | Deposit date: | 2013-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
1BTL
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI TEM1 BETA-LACTAMASE AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | BETA-LACTAMASE TEM1, SULFATE ION | | Authors: | Jelsch, C, Mourey, L, Masson, J.M, Samama, J.P. | | Deposit date: | 1993-11-01 | | Release date: | 1995-01-26 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Escherichia coli TEM1 beta-lactamase at 1.8 A resolution.
Proteins, 16, 1993
|
|
1C0G
 
 | | CRYSTAL STRUCTURE OF 1:1 COMPLEX BETWEEN GELSOLIN SEGMENT 1 AND A DICTYOSTELIUM/TETRAHYMENA CHIMERA ACTIN (MUTANT 228: Q228K/T229A/A230Y/E360H) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, PROTEIN (CHIMERIC ACTIN), ... | | Authors: | Matsuura, Y, Stewart, M, Kawamoto, M, Kamiya, N, Saeki, K, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 1999-07-16 | | Release date: | 2000-03-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the higher Ca(2+)-activation of the regulated actin-activated myosin ATPase observed with Dictyostelium/Tetrahymena actin chimeras.
J.Mol.Biol., 296, 2000
|
|
4NVC
 
 | |
3BNB
 
 | | Lipoxygenase-1 (Soybean) I553L Mutant | | Descriptor: | FE (III) ION, Seed lipoxygenase-1 | | Authors: | Tomchick, D.R. | | Deposit date: | 2007-12-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Enzyme structure and dynamics affect hydrogen tunneling: the impact of a remote side chain (I553) in soybean lipoxygenase-1.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4NVD
 
 | |
4OQ7
 
 | |
4OYI
 
 | | Human solAC Complexed with (4-Amino-furazan-3-yl)-phenyl-methanone | | Descriptor: | (4-azanyl-1,2,5-oxadiazol-3-yl)-phenyl-methanone, Adenylate cyclase type 10, GLYCEROL | | Authors: | Vinkovic, M. | | Deposit date: | 2014-02-12 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human soluble adenylate cyclase reveals a distinct, highly flexible allosteric bicarbonate binding pocket.
Chemmedchem, 9, 2014
|
|
4OYW
 
 | | Crystal Structure of Human Soluble Adenylate Cyclase | | Descriptor: | Adenylate cyclase type 10, CHLORIDE ION, GLYCEROL | | Authors: | Vinkovic, M. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human soluble adenylate cyclase reveals a distinct, highly flexible allosteric bicarbonate binding pocket.
Chemmedchem, 9, 2014
|
|
3BNC
 
 | | Lipoxygenase-1 (Soybean) I553G Mutant | | Descriptor: | ACETIC ACID, FE (III) ION, Seed lipoxygenase-1 | | Authors: | Tomchick, D.R. | | Deposit date: | 2007-12-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enzyme structure and dynamics affect hydrogen tunneling: the impact of a remote side chain (I553) in soybean lipoxygenase-1.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1CZI
 
 | | CHYMOSIN COMPLEX WITH THE INHIBITOR CP-113972 | | Descriptor: | CHYMOSIN, CP-113972 (NORSTATINE-S-METHYL CYSTEINE-IODO-PHENYLALANINE-PROLINE) | | Authors: | Groves, M.R, Dhanaraj, V, Pitts, J.E, Badasso, M, Hoover, D, Nugent, P, Blundell, T.L. | | Deposit date: | 1997-01-15 | | Release date: | 1997-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A 2.3 A resolution structure of chymosin complexed with a reduced bond inhibitor shows that the active site beta-hairpin flap is rearranged when compared with the native crystal structure.
Protein Eng., 11, 1998
|
|
4OPH
 
 | | X-ray structure of full-length H6N6 NS1 | | Descriptor: | Nonstructural protein 1 | | Authors: | Carrillo, B, Choi, J.M, Bornholdt, Z.A, Sankaran, S, Rice, A.P, Prasad, B.V.V. | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.158 Å) | | Cite: | The Influenza A Virus Protein NS1 Displays Structural Polymorphism.
J.Virol., 88, 2014
|
|
3BTO
 
 | |
1P2Y
 
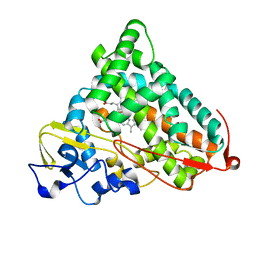 | | CRYSTAL STRUCTURE OF CYTOCHROME P450CAM IN COMPLEX WITH (S)-(-)-NICOTINE | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Cytochrome P450-cam, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Strickler, M, Goldstein, B.M, Maxfield, K, Shireman, L, Kim, G, Matteson, D, Jones, J.P. | | Deposit date: | 2003-04-16 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic Studies on the Complex Behavior of Nicotine Binding to P450cam (CYP101)(dagger).
Biochemistry, 42, 2003
|
|
1DN9
 
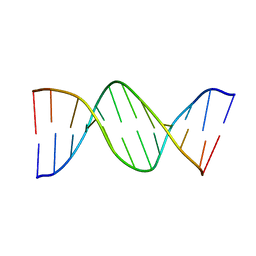 | |
1P7R
 
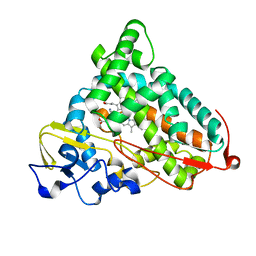 | | CRYSTAL STRUCTURE OF REDUCED, CO-EXPOSED COMPLEX OF CYTOCHROME P450CAM WITH (S)-(-)-NICOTINE | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Cytochrome P450-cam, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Strickler, M, Goldstein, B.M, Maxfield, K, Shireman, L, Kim, G, Matteson, D, Jones, J.P. | | Deposit date: | 2003-05-05 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystallographic Studies on the Complex Behavior of Nicotine Binding to P450cam (CYP101)(dagger).
Biochemistry, 42, 2003
|
|
4OL0
 
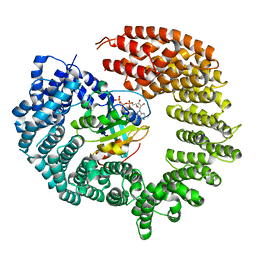 | | Crystal structure of transportin-SR2, a karyopherin involved in human disease, in complex with Ran | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tsirkone, V.G, Strelkov, S.V. | | Deposit date: | 2014-01-23 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of transportin SR2, a karyopherin involved in human disease, in complex with Ran.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3CI5
 
 | |
1MIC
 
 | |
4OR7
 
 | |
1DGZ
 
 | | RIBOSMAL PROTEIN L36 FROM THERMUS THERMOPHILUS: NMR STRUCTURE ENSEMBLE | | Descriptor: | PROTEIN (L36 RIBOSOMAL PROTEIN), ZINC ION | | Authors: | Hard, T, Rak, A, Allard, P, Kloo, L, Garber, M. | | Deposit date: | 1999-11-27 | | Release date: | 1999-12-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ribosomal protein L36 from Thermus thermophilus reveals a zinc-ribbon-like fold.
J.Mol.Biol., 296, 2000
|
|
