2PT2
 
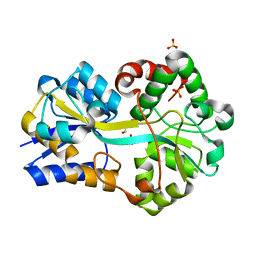 | | Structure of FutA1 with Iron(II) | | Descriptor: | 1,2-ETHANEDIOL, FE (II) ION, Iron transport protein, ... | | Authors: | Koropatkin, N.M, Randich, A.M, Bhattachryya-Pakrasi, M, Pakrasi, H.B, Smith, T.J. | | Deposit date: | 2007-05-07 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the iron-binding protein, FutA1, from Synechocystis 6803.
J.Biol.Chem., 282, 2007
|
|
2PVP
 
 | |
6KLP
 
 | |
2PYK
 
 | |
2PZT
 
 | |
2Q3R
 
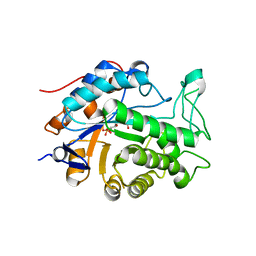 | | Ensemble refinement of the protein crystal structure of At1g76680 from Arabidopsis thaliana | | Descriptor: | 12-oxophytodienoate reductase 1, FLAVIN MONONUCLEOTIDE | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q4F
 
 | | Ensemble refinement of the crystal structure of putative histidine-containing phosphotransfer protein from rice, Ak104879 | | Descriptor: | Histidine-containing phosphotransfer protein 1 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q4M
 
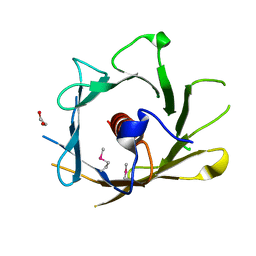 | | Ensemble refinement of the crystal structure of protein from Arabidopsis thaliana At5g01750 | | Descriptor: | 1,2-ETHANEDIOL, Protein At5g01750 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q4W
 
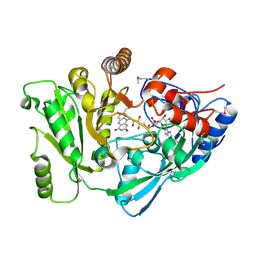 | | Ensemble refinement of the protein crystal structure of cytokinin oxidase/dehydrogenase (CKX) from Arabidopsis thaliana At5g21482 | | Descriptor: | Cytokinin dehydrogenase 7, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Arabidopsis thaliana cytokinin dehydrogenase.
Proteins, 70, 2008
|
|
2Q3T
 
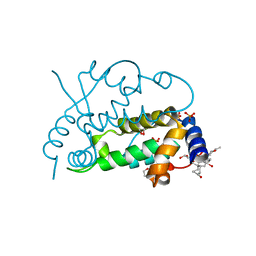 | | Ensemble refinement of the protein crystal structure of gene product from Arabidopsis thaliana At3g22680 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Protein At3g22680, ... | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2PFY
 
 | |
6D3L
 
 | | Crystal structure of unphosphorylated human PKR | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | Erlandsen, H, Mayo, C, Robinson, V.L, Cole, J.L. | | Deposit date: | 2018-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Protein Kinase R Autophosphorylation.
Biochemistry, 58, 2019
|
|
2PUJ
 
 | | Crystal Structure of the S112A/H265A double mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, in complex with its substrate HOPDA | | Descriptor: | (2E,4E)-2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOIC ACID, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, MALONATE ION, ... | | Authors: | Bhowmik, S, Bolin, J.T. | | Deposit date: | 2007-05-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Tautomeric Half-reaction of BphD, a C-C Bond Hydrolase: KINETIC AND STRUCTURAL EVIDENCE SUPPORTING A KEY ROLE FOR HISTIDINE 265 OF THE CATALYTIC TRIAD.
J.Biol.Chem., 282, 2007
|
|
2PWT
 
 | | Crystal structure of the bacterial ribosomal decoding site complexed with aminoglycoside containing the L-HABA group | | Descriptor: | 22-mer of the ribosomal decoding site, DOUBLY FUNCTIONALIZED PAROMOMYCIN PM-II-162 | | Authors: | Kondo, J, Pachamuthu, K, Francois, B, Szychowski, J, Hanessian, S, Westhof, E. | | Deposit date: | 2007-05-13 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Bacterial Ribosomal Decoding Site Complexed with a Synthetic Doubly Functionalized Paromomycin Derivative: a New Specific Binding Mode to an A-Minor Motif Enhances in vitro Antibacterial Activity
Chemmedchem, 2, 2007
|
|
2PW5
 
 | |
1T7X
 
 | | Zn-alpha-2-glycoprotein; refolded CHO-ZAG PEG 400 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Zinc-alpha-2-glycoprotein | | Authors: | Delker, S.L, West Jr, A.P, McDermott, L, Kennedy, M.W, Bjorkman, P.J. | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystallographic studies of ligand binding by Zn-alpha2-glycoprotein.
J.Struct.Biol., 148, 2004
|
|
2PYG
 
 | | Azotobacter vinelandii Mannuronan C-5 epimerase AlgE4 A-module | | Descriptor: | CALCIUM ION, GLYCEROL, Poly(beta-D-mannuronate) C5 epimerase 4 | | Authors: | Rozeboom, H.J, Bjerkan, T.M, Kalk, K.H, Ertesvag, H, Holtan, S, Aachmann, F.L, Valla, S, Dijkstra, B.W. | | Deposit date: | 2007-05-16 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mutational Characterization of the Catalytic A-module of the Mannuronan C-5-epimerase AlgE4 from Azotobacter vinelandii.
J.Biol.Chem., 283, 2008
|
|
2PXH
 
 | | Crystal structure of a bipyridylalanyl-tRNA synthetase | | Descriptor: | 3-(2,2'-BIPYRIDIN-5-YL)-L-ALANINE, Tyrosyl-tRNA synthetase | | Authors: | Liu, W, Xie, J, Schultz, P.G. | | Deposit date: | 2007-05-14 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A genetically encoded bidentate, metal-binding amino acid.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
2Q06
 
 | | Crystal structure of Influenza A Virus H5N1 Nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Ng, A.K.L, Zhang, H, Tan, K, Wang, J, Shaw, P.C. | | Deposit date: | 2007-05-21 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design.
Faseb J., 22, 2008
|
|
2Q2O
 
 | | Crystal structure of H183C Bacillus subtilis ferrochelatase in complex with deuteroporphyrin IX 2,4-disulfonic acid dihydrochloride | | Descriptor: | Ferrochelatase, MAGNESIUM ION, PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-29 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
1T7Z
 
 | | Zn-alpha-2-glycoprotein; baculo-ZAG no PEG, no glycerol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Zinc-alpha-2-glycoprotein | | Authors: | Delker, S.L, West Jr, A.P, McDermott, L, Kennedy, M.W, Bjorkman, P.J. | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies of ligand binding by Zn-alpha2-glycoprotein.
J.Struct.Biol., 148, 2004
|
|
2OM2
 
 | |
2OQB
 
 | |
2OXR
 
 | | PAB0955 crystal structure : a GTPase in GDP and Mg bound form from Pyrococcus abyssi (after GTP hydrolysis) | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
2P14
 
 | | Crystal structure of small subunit (R.BspD6I2) of the heterodimeric restriction endonuclease R.BspD6I | | Descriptor: | GLYCEROL, Heterodimeric restriction endonuclease R.BspD6I small subunit, SULFATE ION | | Authors: | Kachalova, G.S, Bartunik, H.D, Artyukh, R.I, Rogulin, E.A, Yunusova, A.K, Zheleznaya, L.A, Matvienko, N.I. | | Deposit date: | 2007-03-02 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of the heterodimeric type IIS restriction endonuclease R.BspD6I acting as a complex between a monomeric site-specific nickase and a catalytic subunit.
J.Mol.Biol., 384, 2008
|
|
