1GX4
 
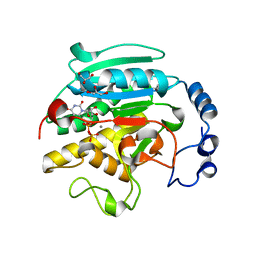 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - N-ACETYL LACTOSAMINE COMPLEX | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-27 | | Release date: | 2003-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
1GWW
 
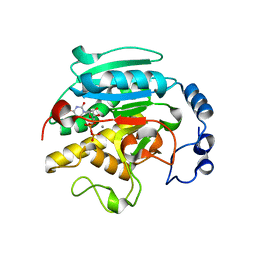 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - ALPHA-D-GLUCOSE COMPLEX | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
7B4O
 
 | |
7B4P
 
 | |
6FTI
 
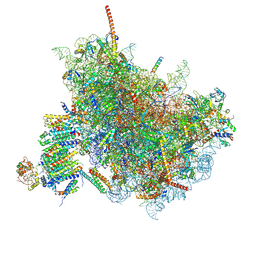 | | Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Programmed 80S Ribosome | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Braunger, K, Becker, T, Beckmann, R. | | Deposit date: | 2018-02-22 | | Release date: | 2018-03-21 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum.
Science, 360, 2018
|
|
1GX0
 
 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - BETA-D-GALACTOSE COMPLEX | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
7SCU
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL7 | | Descriptor: | C-C motif chemokine 7, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-29 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of Tick Evasin EVA-AAM1001 complexed to human chemokine CCL7
To Be Published
|
|
8K2D
 
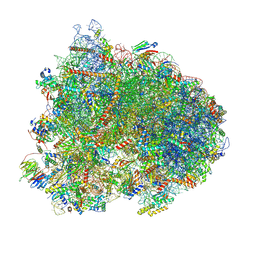 | | Cryo-EM structure of the yeast 80S ribosome with tigecycline, eEF2, Stm1 and eIF5A | | Descriptor: | 18S rRNA, 23S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Beckmann, R, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
7PWF
 
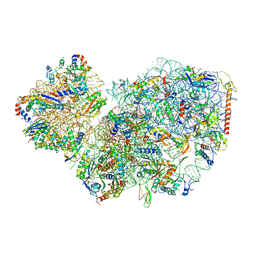 | | Cryo-EM structure of small subunit of Giardia lamblia ribosome at 2.9 A resolution | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | Authors: | Hiregange, D.G, Rivalta, A, Bose, T, Breiner-Goldstein, E, Samiya, S, Cimicata, G, Kulakova, L, Zimmerman, E, Bashan, A, Herzberg, O, Yonath, A. | | Deposit date: | 2021-10-06 | | Release date: | 2022-05-25 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia.
Nucleic Acids Res., 50, 2022
|
|
3JC7
 
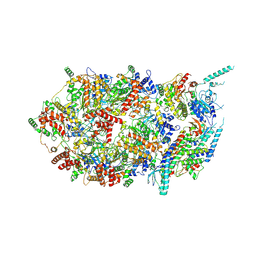 | | Structure of the eukaryotic replicative CMG helicase and pumpjack motion | | Descriptor: | Cell division control protein 45, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Li, H, Bai, L, Yuan, Z, Sun, J, Georgescu, R.E, Liu, J, O'Donnell, M.E. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4LTR
 
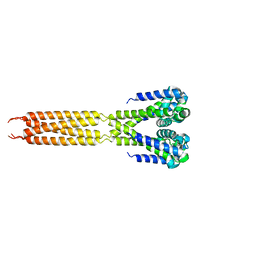 | | Bacterial sodium channel, His245Gly mutant, I222 space group | | Descriptor: | Ion transport protein | | Authors: | Shaya, D, Findeisen, F, Abderemane-Ali, F, Arrigoni, C, Wong, S, Reddy Nurva, S, Loussouarn, G, Minor, D.L. | | Deposit date: | 2013-07-23 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.8 Å) | | Cite: | Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels.
J.Mol.Biol., 426, 2014
|
|
4DH3
 
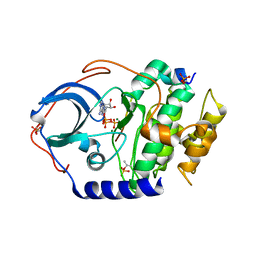 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, ATP and IP20 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4LTQ
 
 | | Bacterial sodium channel in low calcium, P42 space group | | Descriptor: | Ion transport protein | | Authors: | Shaya, D, Findeisen, F, Abderemane-Ali, F, Arrigoni, C, Wong, S, Reddy Nurva, S, Loussouarn, G, Minor, D.L. | | Deposit date: | 2013-07-23 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels.
J.Mol.Biol., 426, 2014
|
|
4DH7
 
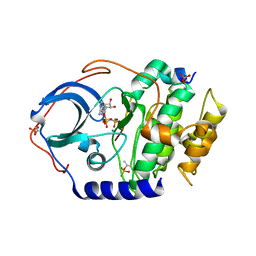 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, AMP-PNP and IP20' | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6PC4
 
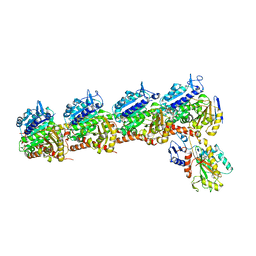 | | Tubulin-RB3_SLD-TTL in complex with compound ABI-274 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2019-06-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structure-Activity Relationship Study of Novel 6-Aryl-2-benzoyl-pyridines as Tubulin Polymerization Inhibitors with Potent Antiproliferative Properties.
J.Med.Chem., 63, 2020
|
|
4DH1
 
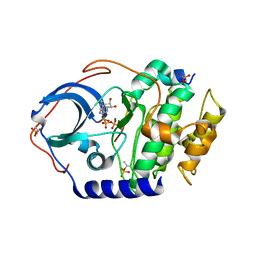 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with low Mg2+, ATP and IP20 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7AEQ
 
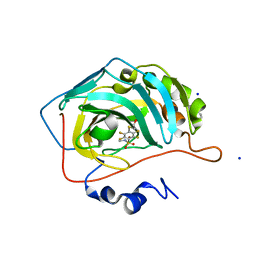 | | Human carbonic anhydrase II in complex with 2,3,5,6-tetrafluoro-4-(2-hydroxyethylsulfanyl)-N-methyl-benzenesulfonamide | | Descriptor: | 2,3,5,6-tetrakis(fluoranyl)-4-(2-hydroxyethylsulfanyl)-~{N}-methyl-benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SODIUM ION, ... | | Authors: | Paketuryte, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and mechanism of secondary sulfonamide binding to carbonic anhydrases.
Eur.Biophys.J., 50, 2021
|
|
7AES
 
 | | Human carbonic anhydrase II in complex with 2,3,5,6-tetrafluoro-N-methyl-4-propylsulfanyl-benzenesulfonamide | | Descriptor: | 2,3,5,6-tetrakis(fluoranyl)-~{N}-methyl-4-propylsulfanyl-benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BICINE, ... | | Authors: | Paketuryte, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and mechanism of secondary sulfonamide binding to carbonic anhydrases.
Eur.Biophys.J., 50, 2021
|
|
7AGN
 
 | | Human carbonic anhydrase II in complex with 4-(2-aminoethylsulfanyl)-2,3,5,6-tetrafluoro-N-methyl-benzenesulfonamide | | Descriptor: | 4-(2-azanylethylsulfanyl)-2,3,5,6-tetrakis(fluoranyl)-~{N}-methyl-benzenesulfonamide, BICINE, SODIUM ION, ... | | Authors: | Paketuryte, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-09-23 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and mechanism of secondary sulfonamide binding to carbonic anhydrases.
Eur.Biophys.J., 50, 2021
|
|
3CVV
 
 | | Drosophila melanogaster (6-4) photolyase bound to ds DNA with a T-T (6-4) photolesion and F0 cofactor | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), ... | | Authors: | Glas, A.F, Maul, M.J, Cryle, M.J, Barends, T.R.M, Schneider, S, Kaya, E, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The archaeal cofactor F0 is a light-harvesting antenna chromophore in eukaryotes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6OD6
 
 | | Structure of BACE-1 in complex with Ligand 13 | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{3-[(3R)-1-amino-3-methyl-3,4-dihydropyrrolo[1,2-a]pyrazin-3-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide | | Authors: | Shaffer, P.L. | | Deposit date: | 2019-03-26 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evaluation of a Series of beta-Secretase 1 Inhibitors Containing Novel Heteroaryl-Fused-Piperazine Amidine Warheads.
Acs Med.Chem.Lett., 10, 2019
|
|
1GXT
 
 | | Hydrogenase Maturation Protein HypF "acylphosphatase-like" N-terminal domain (HypF-ACP) in complex with Sulfate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, HYDROGENASE MATURATION PROTEIN HYPF, ... | | Authors: | Rosano, C, Zuccotti, S, Stefani, M, Bucciantini, M, Ramponi, G, Bolognesi, M. | | Deposit date: | 2002-04-11 | | Release date: | 2002-09-12 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal Structure and Anion Binding in the Prokaryotic Hydrogenase Maturation Factor Hypf Acylphosphatase-Like Domain
J.Mol.Biol., 321, 2002
|
|
1HH7
 
 | |
1GZ6
 
 | | (3R)-HYDROXYACYL-COA DEHYDROGENASE FRAGMENT OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 2 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ESTRADIOL 17 BETA-DEHYDROGENASE 4, SULFATE ION | | Authors: | Haapalainen, A.M, Hiltunen, J.K, Glumoff, T. | | Deposit date: | 2002-05-16 | | Release date: | 2003-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Binary Structure of the Two-Domain (3R)-Hydroxyacyl-Coa Dehydrogenase from Rat Peroxisomal Multifunctional Enzyme Type 2 at 2.38 A Resolution
Structure, 11, 2003
|
|
1H0D
 
 | | Crystal structure of Human Angiogenin in complex with Fab fragment of its monoclonal antibody mAb 26-2F | | Descriptor: | ANGIOGENIN, ANTIBODY FAB FRAGMENT, HEAVY CHAIN, ... | | Authors: | Chavali, G.B, Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 2002-06-19 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Angiogenin in Complex with an Antitumor Neutralizing Antibody
Structure, 11, 2003
|
|
