1QMT
 
 | |
7SCT
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL16 | | Descriptor: | C-C motif chemokine 16, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
7SCV
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL17 | | Descriptor: | C-C motif chemokine 17, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
1H5Q
 
 | | Mannitol dehydrogenase from Agaricus bisporus | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT MANNITOL DEHYDROGENASE, NICKEL (II) ION | | Authors: | Horer, S, Stoop, J, Mooibroek, H, Baumann, U, Sassoon, J. | | Deposit date: | 2001-05-24 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystallographic Structure of the Mannitol 2-Dehydrogenase Nadp+ Binary Complex from Agaricus Bisporus
J.Biol.Chem., 276, 2001
|
|
5CK3
 
 | | Signal recognition particle receptor SRb-GTP/SRX complex from Chaetomium thermophilum | | Descriptor: | GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jadhav, B.R, Wild, K, Sinning, I. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Switch Cycle of SR beta as Ancestral Eukaryotic GTPase Associated with Secretory Membranes.
Structure, 23, 2015
|
|
8AB3
 
 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in complex with oxamate, NADH and FBP. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1,6-di-O-phosphono-beta-D-fructofuranose, L-lactate dehydrogenase, ... | | Authors: | Robin, A.Y, Girard, E, Madern, D. | | Deposit date: | 2022-07-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
8ADB
 
 | | Viral tegument-like DUBs | | Descriptor: | CITRIC ACID, Polyubiquitin-C, Wc-VDT1, ... | | Authors: | Erven, I, Abraham, E.T, Hermanns, T, Baumann, U, Hofmann, K. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A widely distributed family of eukaryotic and bacterial deubiquitinases related to herpesviral large tegument proteins.
Nat Commun, 13, 2022
|
|
8ADD
 
 | | Viral tegument-like DUBs | | Descriptor: | ATP-dependent DNA helicase | | Authors: | Erven, I, Abraham, E.T, Hermanns, T, Hofmann, K, Baumann, U. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A widely distributed family of eukaryotic and bacterial deubiquitinases related to herpesviral large tegument proteins.
Nat Commun, 13, 2022
|
|
8ADC
 
 | | Viral tegument-like DUBs | | Descriptor: | Viral deubiquitinating enzyme | | Authors: | Erven, I, Abraham, E.T, Hermanns, T, Baumann, U, Hofmann, K. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A widely distributed family of eukaryotic and bacterial deubiquitinases related to herpesviral large tegument proteins.
Nat Commun, 13, 2022
|
|
5M43
 
 | |
9FH9
 
 | |
1ZRF
 
 | | 4 crystal structures of CAP-DNA with all base-pair substitutions at position 6, CAP-[6C;17G]ICAP38 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5'-D(*AP*TP*TP*TP*CP*GP*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*TP*CP*GP*AP*AP*AP*T)-3', ... | | Authors: | Berman, H.M, Napoli, A.A. | | Deposit date: | 2005-05-19 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: recognition of pyrimidine-purine and purine-purine steps.
J.Mol.Biol., 357, 2006
|
|
1ZRD
 
 | | 4 crystal structures of CAP-DNA with all base-pair substitutions at position 6, CAP-[6A;17T]ICAP38 DNA | | Descriptor: | 5'-D(*AP*TP*TP*TP*CP*GP*AP*AP*AP*AP*AP*TP*GP*AP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*TP*CP*AP*TP*TP*TP*TP*TP*CP*GP*AP*AP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Berman, H.M, Napoli, A.A. | | Deposit date: | 2005-05-19 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: recognition of pyrimidine-purine and purine-purine steps.
J.Mol.Biol., 357, 2006
|
|
4LX9
 
 | |
4M49
 
 | | Lactate Dehydrogenase A in complex with a substituted pyrazine inhibitor compound 18 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-(5-amino-6-{[(1R)-1-phenylethyl]amino}pyrazin-2-yl)-4-chlorobenzoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2013-08-06 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Identification of 2-amino-5-aryl-pyrazines as inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7C8E
 
 | | Crystal Structure of 14-3-3 epsilon with 9J10 peptide | | Descriptor: | 14-3-3 protein epsilon, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 9J10 | | Authors: | Mathivanan, S, Sudhakar, S, Bairy, S, Kamariah, N, Venkitaraman, A. | | Deposit date: | 2020-05-30 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Target identification for small-molecule discovery in the FOXO3a tumor-suppressor pathway using a biodiverse peptide library.
Cell Chem Biol, 28, 2021
|
|
3EPL
 
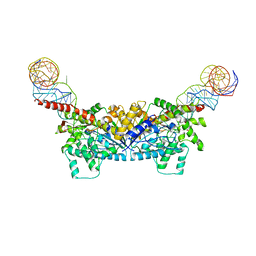 | |
3EPJ
 
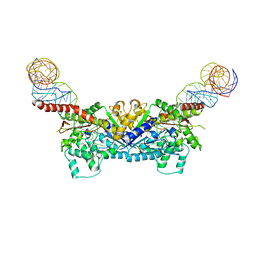 | |
3EPK
 
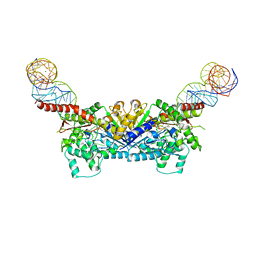 | |
8HWO
 
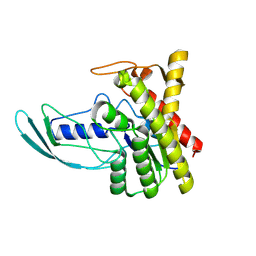 | |
3EPH
 
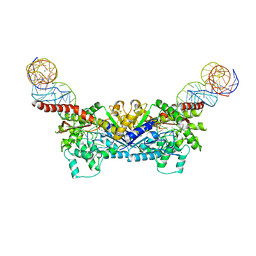 | |
5JJ0
 
 | | Structure of G9a SET-domain with Histone H3K9M peptide and excess SAH | | Descriptor: | Histone H3K9M mutant peptide, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1H64
 
 | |
1GQV
 
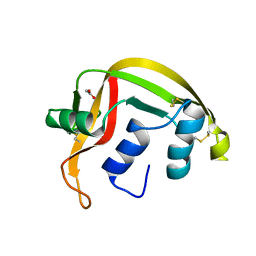 | | Atomic Resolution (0.98A) Structure of Eosinophil-Derived Neurotoxin | | Descriptor: | ACETATE ION, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Swaminathan, G.J, Holloway, D.E, Veluraja, K, Acharya, K.R. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution (0.98 A) Structure of Eosinophil-Derived Neurotoxin
Biochemistry, 41, 2002
|
|
3C63
 
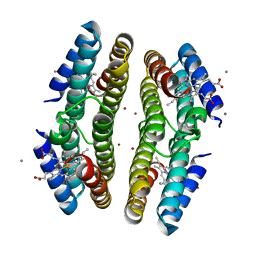 | | Tetrameric Cytochrome cb562 (K34/H59/D62/H63/H73/A74/H77) Assembly Stabilized by Interprotein Zinc Coordination | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562, ... | | Authors: | Tezcan, F.A, Salgado, E.N, Lewis, R.A, Faraone-Mennella, J. | | Deposit date: | 2008-02-02 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Metal-mediated self-assembly of protein superstructures: influence of secondary interactions on protein oligomerization and aggregation.
J.Am.Chem.Soc., 130, 2008
|
|
