4QYY
 
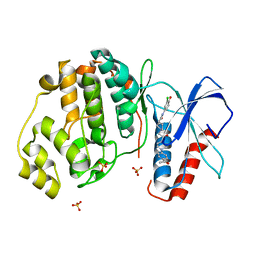 | | Discovery of Novel, Dual Mechanism ERK Inhibitors by Affinity Selection Screening of an Inactive Kinase State | | Descriptor: | (3R)-1-{2-[4-(4-acetylphenyl)piperazin-1-yl]-2-oxoethyl}-N-(3-chloro-4-hydroxyphenyl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Deng, Y, Shipps, G.W, Cooper, A, English, J.M, Annis, D.A, Carr, D, Nan, Y, Wang, T, Zhu, Y.H, Chuang, C, Dayananth, P, Hruza, A.W, Xiao, L, Jin, W, Kirschmeier, P, Windsor, W.T, Samatar, A.A. | | Deposit date: | 2014-07-26 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Novel, Dual Mechanism ERK Inhibitors by Affinity Selection Screening of an Inactive Kinase.
J.Med.Chem., 57, 2014
|
|
7W07
 
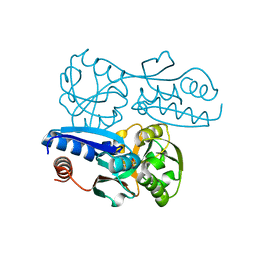 | | Itaconate inducible LysR-Type Transcriptional regulator (ITCR) in complex with itaconate, Space group C121. | | Descriptor: | 2-methylidenebutanedioic acid, SULFATE ION, Transcriptional regulator, ... | | Authors: | Sun, P.K, Wang, B, Li, X.J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A genetically encoded fluorescent biosensor for detecting itaconate with subcellular resolution in living macrophages.
Nat Commun, 13, 2022
|
|
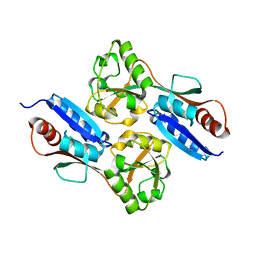
7W08
 
 | |
7W06
 
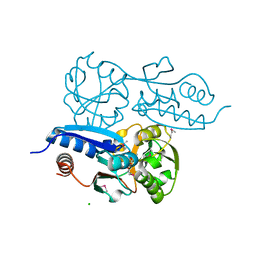 | | Itaconate inducible LysR-Type Transcriptional regulator (ITCR) in complex with itaconate (SeMet labeled), Space group C121. | | Descriptor: | 2-methylidenebutanedioic acid, CHLORIDE ION, SULFATE ION, ... | | Authors: | Sun, P.K, Wang, B, Wang, Z.X, Qi, S, Li, X.J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A genetically encoded fluorescent biosensor for detecting itaconate with subcellular resolution in living macrophages.
Nat Commun, 13, 2022
|
|
6E16
 
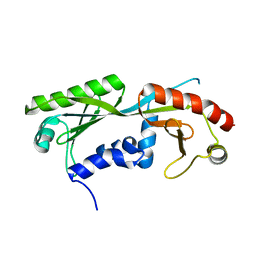 | | Ternary structure of c-Myc-TBP-TAF1 | | Descriptor: | Transcription initiation factor TFIID subunit 1,Myc proto-oncogene protein,TATA-box-binding protein | | Authors: | Wei, Y, Dong, A, Sunnerhagen, M, Penn, L, Tong, Y, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-07-09 | | Release date: | 2019-10-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multiple direct interactions of TBP with the MYC oncoprotein.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8G28
 
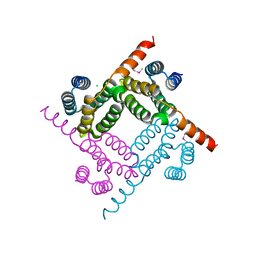 | | Crystal Structure of the C-terminal Fragment of AAA ATPase from Streptococcus pneumoniae. | | Descriptor: | ATPase, AAA family, CHLORIDE ION | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-02-03 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of the C-terminal Fragment of AAA ATPase from Streptococcus pneumoniae.
To Be Published
|
|
8Q7N
 
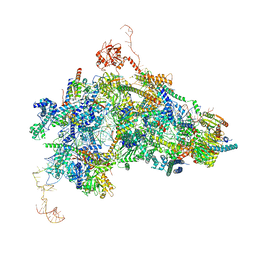 | | cryo-EM structure of the human spliceosomal B complex protomer (tri-snRNP core region) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, MINX pre-mRNA, Microfibrillar-associated protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2023-08-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM analyses of dimerized spliceosomes provide new insights into the functions of B complex proteins.
Embo J., 43, 2024
|
|
7PA2
 
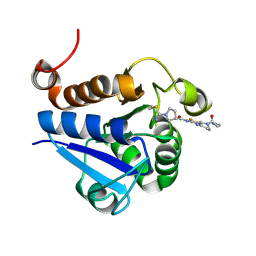 | | PARK7 with inhibitor 8RK64 | | Descriptor: | (3~{S})-~{N}-[5-[2-[(azanylidene-$l^{4}-azanylidene)amino]ethanoyl]-6,7-dihydro-4~{H}-[1,3]thiazolo[5,4-c]pyridin-2-yl]-1-(iminomethyl)pyrrolidine-3-carboxamide, Parkinson disease protein 7 | | Authors: | Kim, R.Q, Jia, Y, Sapmaz, A, Geurink, P.P. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Chemical Toolkit for PARK7: Potent, Selective, and High-Throughput.
J.Med.Chem., 65, 2022
|
|
7PA3
 
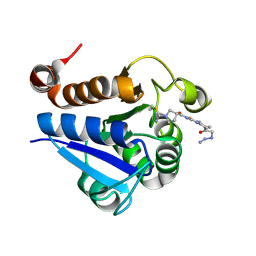 | | PARK7 with covalent inhibitor JYQ-88 | | Descriptor: | (3~{S})-~{N}-[5-[2-[(azanylidene-$l^{4}-azanylidene)amino]ethanoyl]-6,7-dihydro-4~{H}-[1,3]thiazolo[5,4-c]pyridin-2-yl]-1-(iminomethyl)pyrrolidine-3-carboxamide, Parkinson disease protein 7 | | Authors: | Kim, R.Q, Jia, Y, Sapmaz, A, Geurink, P.P. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Chemical Toolkit for PARK7: Potent, Selective, and High-Throughput.
J.Med.Chem., 65, 2022
|
|
2MGQ
 
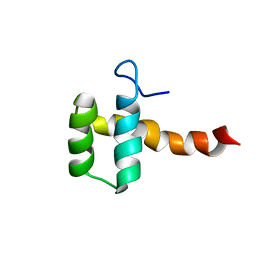 | | Structure of CEH37 Homeodomain | | Descriptor: | Homeobox protein ceh-37 | | Authors: | Moon, S, Lee, W. | | Deposit date: | 2013-11-04 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of CEH37 Homeodomain
To be Published
|
|
6YLE
 
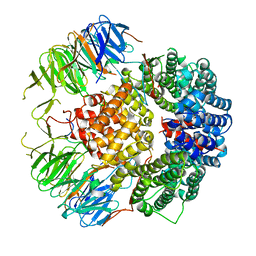 | |
2M34
 
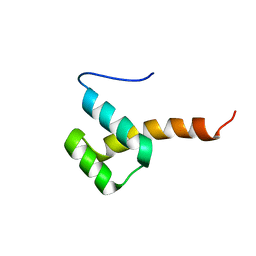 | |
2C6O
 
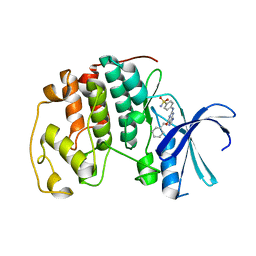 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, O6-CYCLOHEXYLMETHOXY-2-(4'-SULPHAMOYLANILINO) PURINE | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-10 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Triazolo[1,5-a]pyrimidines as novel CDK2 inhibitors: protein structure-guided design and SAR.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
4RUJ
 
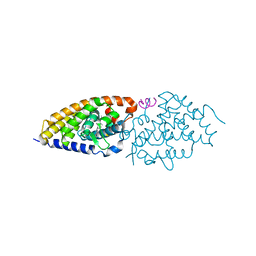 | | Crystal structure of zVDR L337H mutant-VD complex | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Huet, T, Moras, D, Rochel, N. | | Deposit date: | 2014-11-20 | | Release date: | 2015-10-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | A vitamin D receptor selectively activated by gemini analogs reveals ligand dependent and independent effects.
Cell Rep, 10, 2015
|
|
2CCH
 
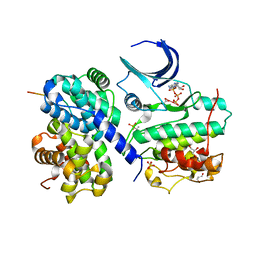 | | The crystal structure of CDK2 cyclin A in complex with a substrate peptide derived from CDC modified with a gamma-linked ATP analogue | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION CONTROL PROTEIN 6 HOMOLOG, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Role of the Phospho-Cdk2/Cyclin a Recruitment Site in Substrate Recognition
J.Biol.Chem., 281, 2006
|
|
2C6T
 
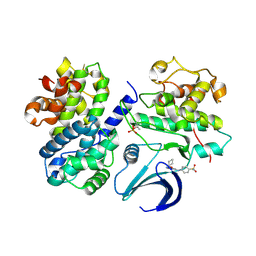 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | 4-{[5-(CYCLOHEXYLOXY)[1,2,4]TRIAZOLO[1,5-A]PYRIMIDIN-7-YL]AMINO}BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-11 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Triazolo[1,5-A]Pyrimidines as Novel Cdk2 Inhibitors: Protein Structure-Guided Design and Sar.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4S2Z
 
 | |
2CLX
 
 | | 4-Arylazo-3,5-diamino-1H-pyrazole CDK Inhibitors: SAR Study, Crystal Structure in Complex with CDK2, Selectivity, and Cellular Effects | | Descriptor: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Krystof, V, Cankar, P, Frysova, I, Slouka, J, Kontopidis, G, Dzubak, P, Hajduch, M, Deazevedo, W.F, Paprskarova, M, Orsag, M, Rolcik, J, Latr, A, Fischer, P.M, Strnad, M. | | Deposit date: | 2006-05-02 | | Release date: | 2006-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 4-Arylazo-3,5-Diamino-1H-Pyrazole Cdk Inhibitors: Sar Study, Crystal Structure in Complex with Cdk2, Selectivity, and Cellular Effects
J.Med.Chem., 49, 2006
|
|
8SOS
 
 | |
4S34
 
 | | ERK2 (I84A) in complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Livnah, O, Karamansha, Y, Tzarum, N. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multiple mechanisms render Erk proteins MEK-independent
To be Published
|
|
4S32
 
 | |
8HJB
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA with coenzyme A | | Descriptor: | COENZYME A, TetR family transcriptional regulator | | Authors: | Liang, H, Bartlam, M. | | Deposit date: | 2022-11-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
4S30
 
 | |
8CSG
 
 | | Human PRMT5:MEP50 structure with Fragment 1 and MTA Bound | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, 6-bromo-1H-pyrrolo[3,2-b]pyridin-5-amine, ... | | Authors: | Gunn, R.J, Lawson, J.D, Smith, C.R. | | Deposit date: | 2022-05-12 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
8CTB
 
 | | Human PRMT5:MEP50 structure with Fragment 3 and MTA Bound | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 7-chloro-1-methyl-1H-benzimidazol-2-amine, Methylosome protein 50, ... | | Authors: | Gunn, R.J, Lawson, J.D, Smith, C.R. | | Deposit date: | 2022-05-13 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
