6IZ4
 
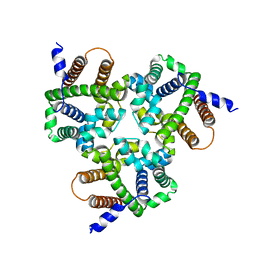 | | Crystal Structure Analysis of TRIC counter-ion channels in calcium release | | Descriptor: | Trimeric intracellular cation channel type B-B | | Authors: | Wang, X.H, Zeng, Y, Gao, F, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8A0Y
 
 | |
7R84
 
 | |
6DAY
 
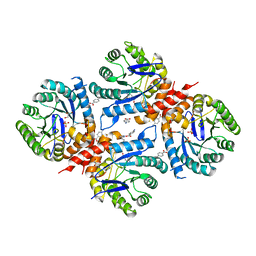 | | Xanthomonas albilineans Dihydropteroate synthase with 4-aminobenzoic acid at 1.65 A | | Descriptor: | 4-AMINOBENZOIC ACID, Dihydropteroate synthase, GLYCEROL, ... | | Authors: | Oliveira, A.A, Guido, R.V.C, Lima, G.M.A, Bueno, R.V, Maluf, F.V. | | Deposit date: | 2018-05-02 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Xanthomonas albilineans Dihydropteroate synthase with 4-aminobenzoic acid
To Be Published
|
|
7ZRR
 
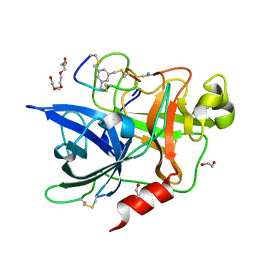 | | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK965 | | Descriptor: | 1,2-ETHANEDIOL, 1,3,5-tris(bromomethyl)benzene, AMINO GROUP, ... | | Authors: | Caregnato, A, Angela, P, Mazzoccato, Y, Frasson, N, Angelini, A, Cendron, L. | | Deposit date: | 2022-05-05 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK965
To Be Published
|
|
8SIP
 
 | |
7AH1
 
 | | L19 diabody fragment from immunocytokine L19-IL2 | | Descriptor: | Anti-(ED-B) scFV | | Authors: | Ongaro, T, Guarino, S.R, Scietti, L, Palamini, M, Wulhfard, S, Villa, A, Neri, D, Forneris, F. | | Deposit date: | 2020-09-23 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inference of molecular structure for characterization and improvement of clinical grade immunocytokines.
J.Struct.Biol., 213, 2021
|
|
6J0O
 
 | | Crystal structure of CERT START domain in complex with compound SC1 | | Descriptor: | 2-[4-[2-fluoranyl-5-[3-(6-methylpyridin-2-yl)-1~{H}-pyrazol-4-yl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT, UNKNOWN ATOM OR ION | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-12-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
8SUD
 
 | | JC Polyomavirus LTA NLS bound to importin alpha 2 | | Descriptor: | Importin subunit alpha-1, Large T antigen | | Authors: | Cross, E.M, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-05-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin alpha-dependent nuclear localization signals.
Protein Sci., 33, 2024
|
|
6J0X
 
 | | Crystal Structure of Yeast Rtt107 and Mms22 | | Descriptor: | Peptide from E3 ubiquitin-protein ligase substrate receptor MMS22, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
7B0T
 
 | | Crystal structure of MLLT1 YEATS domain T3 mutant in complex with benzimidazole-amide based compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 3-iodanyl-4-methyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Ni, X, Chaikuad, A, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-21 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and Inhibitor Binding Characterization of Oncogenic MLLT1 Mutants.
Acs Chem.Biol., 16, 2021
|
|
7ZYW
 
 | | Crystal structure of T2R-TTL-PM534 complex | | Descriptor: | (4R)-N-[(1R)-1-[4-(cyclopropylmethoxy)-6-oxidanylidene-pyran-2-yl]butyl]-4-methyl-2-[(E)-C-methyl-N-oxidanyl-carbonimidoyl]-5H-1,3-thiazole-4-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Oliva, M.A, Diaz, J.F, Cuevas, C. | | Deposit date: | 2022-05-25 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | PM534, an Optimized Target-Protein Interaction Strategy through the Colchicine Site of Tubulin.
J.Med.Chem., 67, 2024
|
|
6J5B
 
 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*GP*GP*TP*AP*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*CP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*TP*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*GP*TP*AP*CP*C)-3'), Protein PHOSPHATE STARVATION RESPONSE 1 | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N, Wu, Y.K. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
7AM3
 
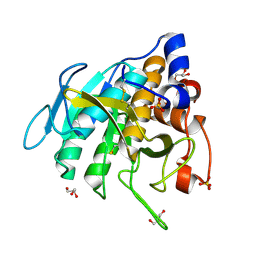 | | Crystal structure of Peptiligase mutant - M222P | | Descriptor: | GLYCEROL, SULFATE ION, Subtilisin BPN' | | Authors: | Rozeboom, H.J, Janssen, D.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | From thiol-subtilisin to omniligase: Design and structure of a broadly applicable peptide ligase.
Comput Struct Biotechnol J, 19, 2021
|
|
7ZRT
 
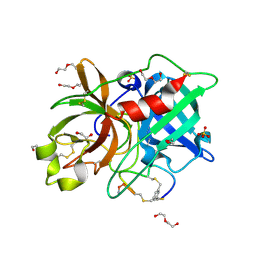 | | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK970 | | Descriptor: | 1,2-ETHANEDIOL, 1,3,5-tris(bromomethyl)benzene, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Caregnato, A, Angela, P, Mazzoccato, Y, Frasson, N, Angelini, A, Cendron, L. | | Deposit date: | 2022-05-05 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK970
To Be Published
|
|
6DCZ
 
 | |
6JAH
 
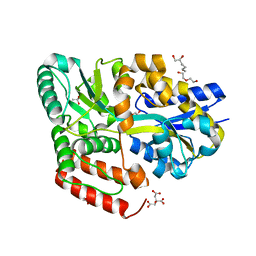 | | Crystal structure of ABC transporter alpha-glycoside-binding protein in complex with glucose | | Descriptor: | (2S)-heptane-1,2,7-triol, 1,2-ETHANEDIOL, ABC transporter, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
6DD4
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropyl-dihydropteridine inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-5,8-dipropyl-7,8-dihydropteridin-6(5H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | dos Reis, C.V, Santiago, A.S, de Souza, G.P, Counago, R.M, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-05-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropyl-dihydropteridine inhibitor
To Be Published
|
|
6D89
 
 | |
8A42
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AmiA in complex with an unknown peptide | | Descriptor: | Oligopeptide-binding protein AmiA, Unknown peptide | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2022-06-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
6JBB
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein W287A in complex with sucrose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-25 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
7B10
 
 | | Crystal structure of MLLT1 YEATS domain T1 mutant in complex with benzimidazole-amide based compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 3-iodanyl-4-methyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, IODIDE ION, ... | | Authors: | Chaikuad, A, Ni, X, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure and Inhibitor Binding Characterization of Oncogenic MLLT1 Mutants.
Acs Chem.Biol., 16, 2021
|
|
8A52
 
 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c-axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Putative Sensory box protein,Putative Sensory box protein,Sensor protein FixL | | Authors: | Batra-Safferling, R, Arinkin, V, Granzin, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c axis)
To Be Published
|
|
7RMZ
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-63 | | Descriptor: | 3C-like proteinase, 6-{4-[3-chloro-4-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
6D96
 
 | |
