6HCP
 
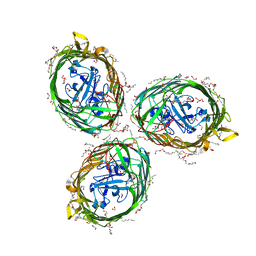 | | Crystal structure of BauA, the Ferric preacinetobactin receptor from Acinetobacter baumannii | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, BauA, ... | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Preacinetobactin not acinetobactin is essential for iron uptake by the BauA transporter of the pathogenAcinetobacter baumannii.
Elife, 7, 2018
|
|
5KLI
 
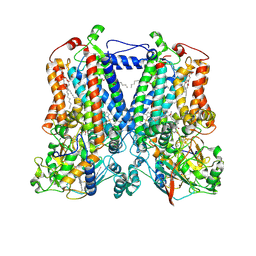 | | Rhodobacter sphaeroides bc1 with stigmatellin and antimycin | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, (2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE, Cytochrome b, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Tang, W.K, Yu, C.A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.996 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
5KLV
 
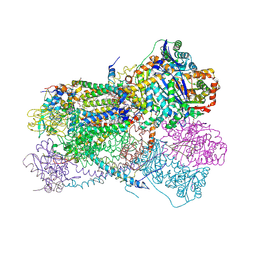 | | Structure of bos taurus cytochrome bc1 with fenamidone inhibited | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, (5S)-5-methyl-2-(methylsulfanyl)-5-phenyl-3-(phenylamino)-3,5-dihydro-4H-imidazol-4-one, 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Zhou, Y, Xiao, Y, Tang, W.K, Yu, C.A, Qin, Z. | | Deposit date: | 2016-06-25 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
6H7F
 
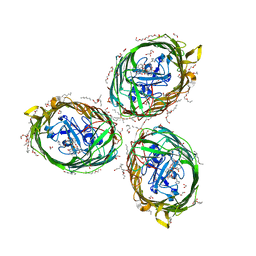 | | Crystal structure of BauA, the Ferric preacinetobactin receptor from Acinetobacter baumannii in complex with Fe3+-Preacinetobactin-acinetobactin | | Descriptor: | (4~{S},5~{R})-2-[2,3-bis(oxidanyl)phenyl]-~{N}-[2-(1~{H}-imidazol-4-yl)ethyl]-5-methyl-~{N}-oxidanyl-4,5-dihydro-1,3-oxazole-4-carboxamide, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, ... | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Preacinetobactin not acinetobactin is essential for iron uptake by the BauA transporter of the pathogenAcinetobacter baumannii.
Elife, 7, 2018
|
|
6S7D
 
 | | Self-complementary duplex DNA containing an internucleoside phosphoroselenolate | | Descriptor: | BARIUM ION, CHLORIDE ION, DNA (5'-D(*GP*(XCI)P*CP*CP*CP*GP*GP*GP*AP*C)-3'), ... | | Authors: | Conlon, P.F, Steinhogl, J, Vyle, J.S, Hall, J.P. | | Deposit date: | 2019-07-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Solid-phase synthesis and structural characterisation of phosphoroselenolate-modified DNA: a backbone analogue which does not impose conformational bias and facilitates SAD X-ray crystallography.
Chem Sci, 10, 2019
|
|
5KKZ
 
 | | Rhodobacter sphaeroides bc1 with famoxadone | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, ASCORBIC ACID, Cytochrome b, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Tang, W.K, Yu, C.A. | | Deposit date: | 2016-06-23 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
4I0Z
 
 | | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates | | Descriptor: | 2-{(1S)-1-[(6-CHLORO-3,3-DIMETHYL-3,4-DIHYDROISOQUINOLIN-1-YL)AMINO]-2-PHENYLETHYL}-4-OXO-1,4-DIHYDROPYRIMIDINE-5-CARBONITRILE, 2-{(1S)-1-[(6-chloro-3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)amino]-2-phenylethyl}-4-oxo-1,4-dihydropyrimidine-5-carbonitrile, ZINC ION | | Authors: | Lougheed, J.C, Brecht, E, Yao, N.H. | | Deposit date: | 2012-11-19 | | Release date: | 2013-03-06 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HZT
 
 | | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates | | Descriptor: | 3-{(1S)-1-[(6-chloro-3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)amino]-2-phenylethyl}-1,2,4-oxadiazol-5(2H)-one, Beta-secretase 1, ZINC ION | | Authors: | Yao, N, Brecht, E. | | Deposit date: | 2012-11-15 | | Release date: | 2013-03-06 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5KY7
 
 | | mouse POFUT1 in complex with O-glucosylated EGF(+) and GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EGF(+), GDP-fucose protein O-fucosyltransferase 1, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-21 | | Release date: | 2017-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
5BJ3
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE TETRA MUTANT 1 | | Descriptor: | PROTEIN (ASPARTATE AMINOTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Kawaguchi, S.I, Miyahara, I, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-01-11 | | Release date: | 2003-09-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme
J.BIOCHEM.(TOKYO), 130, 2001
|
|
6SO3
 
 | | The interacting head motif in insect flight muscle myosin thick filaments | | Descriptor: | Myosin 2 essential light chain striated muscle, Myosin 2 heavy chain striated muscle, Myosin 2 regulatory light chain striated muscle | | Authors: | Morris, E.P, Knupp, C, Squire, J.M. | | Deposit date: | 2019-08-28 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | The Interacting Head Motif Structure Does Not Explain the X-Ray Diffraction Patterns in Relaxed Vertebrate (Bony Fish) Skeletal Muscle and Insect (Lethocerus) Flight Muscle.
Biology (Basel), 8, 2019
|
|
5F87
 
 | |
6U31
 
 | | The crystal structure of 4-(1H-imidazol-1-yl)benzoate-bound CYP199A4 | | Descriptor: | 4-(1H-imidazol-1-yl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | To Be, or Not to Be, an Inhibitor: A Comparison of Azole Interactions with and Oxidation by a Cytochrome P450 Enzyme.
Inorg.Chem., 61, 2022
|
|
5F84
 
 | |
6ZBX
 
 | | Structure of the catalytic domain of the Bacillus circulans alpha-1,6 Mannanase in complex with an alpha-1,6- alpha-manno-cyclophellitol carbasugar-stabilised trisaccharide inhibitor | | Descriptor: | (1~{S},2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, (1~{S},4~{S},5~{R})-6-(hydroxymethyl)cyclohexane-1,2,3,4,5-pentol, 1,2-ETHANEDIOL, ... | | Authors: | Schroeder, S, Offen, W.A, Jin, Y, De Boer, C, Enoterpi, J, Marino, L, van der Marel, G.A, Codee, J.D.C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2020-06-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Development of Non-Hydrolysable Oligosaccharide Activity-Based Inactivators for Endoglycanases: A Case Study on alpha-1,6 Mannanases.
Chemistry, 27, 2021
|
|
6OR2
 
 | | MmpL3 is a lipid transporter that binds trehalose monomycolate and phosphatidylethanolamine | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, DODECYL-BETA-D-MALTOSIDE, Membrane protein, ... | | Authors: | Su, C.-C. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | MmpL3 is a lipid transporter that binds trehalose monomycolate and phosphatidylethanolamine.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5F85
 
 | |
5GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, GLUTATHIONYLSPERMIDINE COMPLEX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, GLUTATHIONYLSPERMIDINE DISULFIDE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
1JLT
 
 | | Vipoxin Complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHOLIPASE A2, ... | | Authors: | Banumathi, S, Rajashankar, K.R, Notzel, C, Aleksiev, B, Singh, T.P, Genov, N, Betzel, C. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the neurotoxic complex vipoxin at 1.4 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5F86
 
 | |
6VER
 
 | | Human insulin analog: [GluB10,TyrB20]-DOI | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | Deposit date: | 2020-01-02 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.047 Å) | | Cite: | Mini-Ins: A minimal, bioactive insulin analog with alternative binding modes
not published
|
|
4ZL6
 
 | |
4ZL5
 
 | |
4ZLW
 
 | |
4ZKW
 
 | |
