3IJ1
 
 | | Crystal structure of Eed in complex with a trimethylated histone H4K20 peptide | | Descriptor: | Histone H4K20 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
3IAG
 
 | | CSL (RBP-Jk) bound to HES-1 nonconsensus site | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*AP*CP*AP*CP*GP*AP*T)-3', 5'-D(*TP*TP*AP*TP*CP*GP*TP*GP*TP*GP*AP*AP*AP*GP*A)-3', ... | | Authors: | Friedmann, D.R, Kovall, R.A. | | Deposit date: | 2009-07-13 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural insights into CSL-DNA complexes.
Protein Sci., 19, 2010
|
|
1ZDI
 
 | | RNA BACTERIOPHAGE MS2 COAT PROTEIN/RNA COMPLEX | | Descriptor: | PROTEIN (RNA BACTERIOPHAGE MS2 COAT PROTEIN), RNA (5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*UP*AP*CP*CP*CP*AP*U P*GP*U)-3') | | Authors: | Valegard, K, Van Den Worm, S, Liljas, L. | | Deposit date: | 1996-09-24 | | Release date: | 1997-04-21 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions.
J.Mol.Biol., 270, 1997
|
|
8R5H
 
 | | Ubiquitin ligation to neosubstrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB-BRD4 BD2 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 5-azanylpentan-2-one, Bromodomain-containing protein 4, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-11-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
1YO7
 
 | |
1A53
 
 | | COMPLEX OF INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS WITH INDOLE-3-GLYCEROLPHOSPHATE AT 2.0 A RESOLUTION | | Descriptor: | INDOLE-3-GLYCEROL PHOSPHATE, INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE | | Authors: | Hennig, M, Darimont, B, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1998-02-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The catalytic mechanism of indole-3-glycerol phosphate synthase: crystal structures of complexes of the enzyme from Sulfolobus solfataricus with substrate analogue, substrate, and product.
J.Mol.Biol., 319, 2002
|
|
1AN7
 
 | |
2B1C
 
 | |
2XL4
 
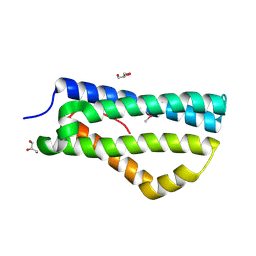 | | LntA, a virulence factor from Listeria monocytogenes | | Descriptor: | GLYCEROL, Listeria nuclear targeted protein A | | Authors: | Lebreton, A, Job, V, Tham, T.N, Camejo, A, Mattei, P.J, Regnault, B, Cabanes, D, Dessen, A, Cossart, P, Bierne, H. | | Deposit date: | 2010-07-19 | | Release date: | 2011-02-02 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response.
Science, 331, 2011
|
|
1ZDH
 
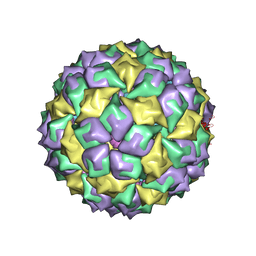 | | MS2 COAT PROTEIN/RNA COMPLEX | | Descriptor: | PROTEIN (BACTERIOPHAGE MS2 COAT PROTEIN), RNA (5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*CP*AP*CP*CP*CP*AP*U P*GP*U)-3') | | Authors: | Valegard, K, Van Den Worm, S, Liljas, L. | | Deposit date: | 1996-09-24 | | Release date: | 1997-04-21 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions.
J.Mol.Biol., 270, 1997
|
|
1Z9C
 
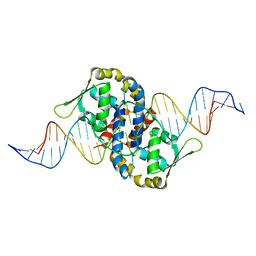 | | Crystal structure of OhrR bound to the ohrA promoter: Structure of MarR family protein with operator DNA | | Descriptor: | DNA (29-MER), Organic hydroperoxide resistance transcriptional regulator | | Authors: | Hong, M, Fuangthong, M, Helmann, J.D, Brennan, R.G. | | Deposit date: | 2005-04-01 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of an OhrR-ohrA Operator Complex Reveals the DNA Binding Mechanism of the MarR Family.
Mol.Cell, 20, 2005
|
|
2B1B
 
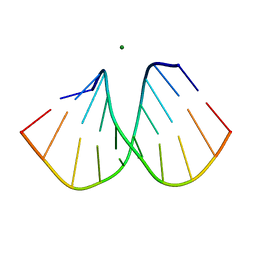 | |
7KTR
 
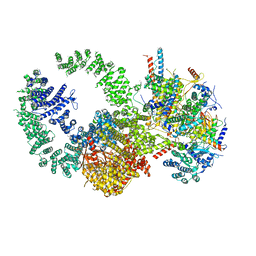 | | Cryo-EM structure of the human SAGA coactivator complex (TRRAP, core) | | Descriptor: | Ataxin-7, INOSITOL HEXAKISPHOSPHATE, Isoform 3 of Transcription factor SPT20 homolog, ... | | Authors: | Herbst, D.A, Esbin, M.N, Nogales, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KTS
 
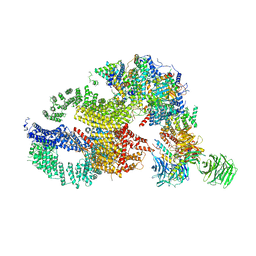 | | Negative stain EM structure of the human SAGA coactivator complex (TRRAP, core, splicing module) | | Descriptor: | Ataxin-7, Isoform 3 of Transcription factor SPT20 homolog, STAGA complex 65 subunit gamma, ... | | Authors: | Herbst, D.A, Esbin, M.N, Nogales, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (19.09 Å) | | Cite: | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2X6U
 
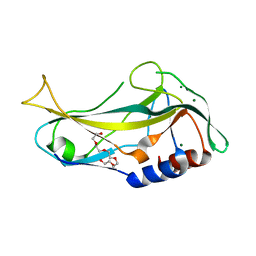 | | Crystal structure of human TBX5 in the DNA-free form | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, MAGNESIUM ION, T-BOX TRANSCRIPTION FACTOR TBX5 | | Authors: | Stirnimann, C.U, Mueller, C.W. | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Tbx5-DNA Recognition: The T-Box Domain in its DNA-Bound and -Unbound Form.
J.Mol.Biol., 400, 2010
|
|
2WVB
 
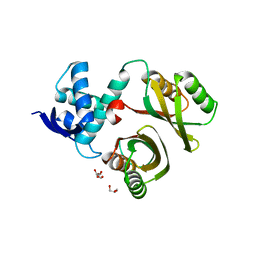 | | Structural and mechanistic insights into Helicobacter pylori NikR function | | Descriptor: | FORMIC ACID, GLYCEROL, PUTATIVE NICKEL-RESPONSIVE REGULATOR | | Authors: | Dian, C, Bahlawane, C, Muller, C, Round, A, Delay, C, Fauquant, C, Schauer, K, De Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2009-10-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insights Into Helicobacter Pylori Nikr Activation.
Nucleic Acids Res., 38, 2010
|
|
2WVF
 
 | | Structural and mechanistic insights into Helicobacter pylori NikR function | | Descriptor: | FORMIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Dian, C, Bahlawane, C, Muller, C, Round, A, Delay, C, Fauquant, C, Schauer, K, de Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2009-10-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Mechanistic Insights Into Helicobacter Pylori Nikr Activation.
Nucleic Acids Res., 38, 2010
|
|
7ENJ
 
 | | Human Mediator (deletion of MED1-IDR) in a Tail-bent conformation (MED-B) | | Descriptor: | Isoform 2 of Mediator of RNA polymerase II transcription subunit 8, Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, ... | | Authors: | Yin, X, Li, J, Wu, Z, Liu, W, Xu, Y. | | Deposit date: | 2021-04-17 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7ENA
 
 | | TFIID-based PIC-Mediator holo-complex in pre-assembled state (pre-hPIC-MED) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Wu, Z, Yin, X, Li, J, Liu, W, Xu, Y. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7ENC
 
 | | TFIID-based PIC-Mediator holo-complex in fully-assembled state (hPIC-MED) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Wu, Z, Yin, X, Li, J, Liu, W, Xu, Y. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
2WVD
 
 | | Structural and mechanistic insights into Helicobacter pylori NikR function | | Descriptor: | GLYCEROL, PUTATIVE NICKEL-RESPONSIVE REGULATOR, SULFATE ION | | Authors: | Dian, C, Bahlawane, C, Muller, C, Round, A, Delay, C, Fauquant, C, Schauer, K, de Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2009-10-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Mechanistic Insights Into Helicobacter Pylori Nikr Activation.
Nucleic Acids Res., 38, 2010
|
|
2WVE
 
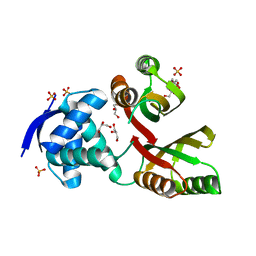 | | Structural and mechanistic insights into Helicobacter pylori NikR function | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRIC ACID, GLYCEROL, ... | | Authors: | Dian, C, Bahlawane, C, Muller, C, Round, A, Delay, C, Fauquant, C, Schauer, K, de Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2009-10-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights Into Helicobacter Pylori Nikr Activation.
Nucleic Acids Res., 38, 2010
|
|
1LS3
 
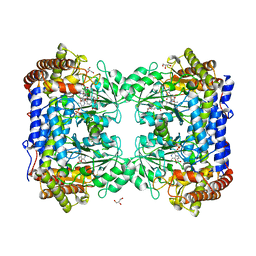 | | Crystal Structure of the Complex between Rabbit Cytosolic Serine Hydroxymethyltransferase and TriGlu-5-formyl-tetrahydrofolate | | Descriptor: | 2-[4-(4-{4-[(2-AMINO-5-FORMYL-4-OXO-3,4,5,6,7,8-HEXAHYDRO-PTERIDIN-6-YLMETHYL)-AMINO]-BENZOYLAMINO}-4-CARBOXY-BUTYRYLAM INO)-4-CARBOXY-BUTYRYLAMINO]-PENTANEDIOIC ACID, GLYCEROL, GLYCINE, ... | | Authors: | Fu, T.F, Scarsdale, J.N, Kazanina, G, Schirch, V, Wright, H.T. | | Deposit date: | 2002-05-16 | | Release date: | 2003-02-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Location of the Pteroylpolyglutamate Binding Site on Rabbit Cytosolic Serine Hydroxymethyltransferase
J.Biol.Chem., 278, 2003
|
|
1LE8
 
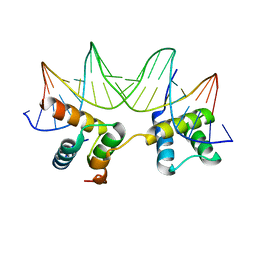 | | Crystal Structure of the MATa1/MATalpha2-3A Heterodimer Bound to DNA Complex | | Descriptor: | 5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*AP*AP*AP*TP*TP*TP*AP*CP*AP*TP*CP*A)-3', 5'-D(*TP*TP*GP*AP*TP*GP*TP*AP*AP*AP*TP*TP*TP*TP*TP*AP*CP*AP*TP*G)-3', MATING-TYPE PROTEIN A-1, ... | | Authors: | Ke, A, Mathias, J.R, Vershon, A.K, Wolberger, C. | | Deposit date: | 2002-04-09 | | Release date: | 2002-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Thermodynamic Characterization of the DNA Binding Properties of a Triple Alanine Mutant of MATalpha2
Structure, 10, 2002
|
|
7EMF
 
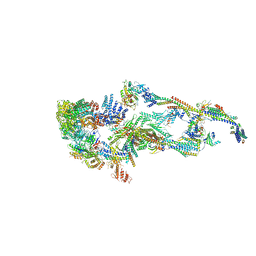 | | Human Mediator (deletion of MED1-IDR) in a Tail-extended conformation | | Descriptor: | Isoform 2 of Mediator of RNA polymerase II transcription subunit 16, Isoform 2 of Mediator of RNA polymerase II transcription subunit 8, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Yin, X, Li, J, Wu, Z, Liu, W, Xu, Y. | | Deposit date: | 2021-04-13 | | Release date: | 2021-05-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
