1N23
 
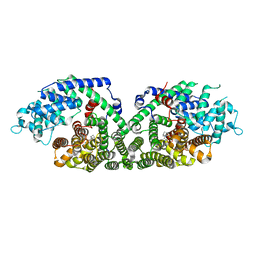 | | (+)-Bornyl diphosphate synthase: Complex with Mg, pyrophosphate, and (1R,4S)-2-azabornane | | Descriptor: | (+)-bornyl diphosphate synthase, (1R,4S)-2-AZABORNANE, MAGNESIUM ION, ... | | Authors: | Whittington, D.A, Wise, M.L, Urbansky, M, Coates, R.M, Croteau, R.B, Christianson, D.W. | | Deposit date: | 2002-10-21 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bornyl diphosphate synthase: Structure and strategy for carbocation manipulation by a terpenoid cyclase
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N24
 
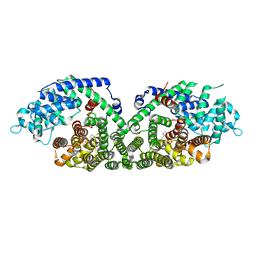 | | (+)-Bornyl diphosphate synthase: Complex with Mg and product | | Descriptor: | (+)-BORNYL DIPHOSPHATE, (+)-bornyl diphosphate synthase, MAGNESIUM ION | | Authors: | Whittington, D.A, Wise, M.L, Urbansky, M, Coates, R.M, Croteau, R.B, Christianson, D.W. | | Deposit date: | 2002-10-21 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bornyl diphosphate synthase: Structure and strategy for carbocation manipulation by a terpenoid cyclase
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5WUA
 
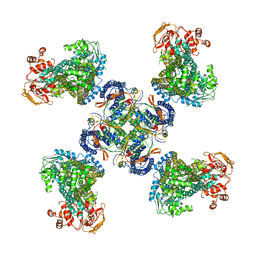 | | Structure of a Pancreatic ATP-sensitive Potassium Channel | | Descriptor: | ATP-sensitive inward rectifier potassium channel 11,superfolder GFP, SUR1 | | Authors: | Li, N, Wu, J.-X, Chen, L, Gao, N. | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure of a Pancreatic ATP-Sensitive Potassium Channel
Cell, 168, 2017
|
|
5AZA
 
 | |
1MV3
 
 | |
4ATZ
 
 | |
1N20
 
 | | (+)-Bornyl Diphosphate Synthase: Complex with Mg and 3-aza-2,3-dihydrogeranyl diphosphate | | Descriptor: | (+)-bornyl diphosphate synthase, 2-[METHYL-(4-METHYL-PENT-3-ENYL)-AMINO]-ETHYL-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Whittington, D.A, Wise, M.L, Urbansky, M, Coates, R.M, Croteau, R.B, Christianson, D.W. | | Deposit date: | 2002-10-21 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bornyl Diphosphate Synthase: Structure and Strategy for Carbocation Manipulation by a Terpenoid Cyclase
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MUZ
 
 | |
3C13
 
 | | Low pH-value crystal structure of emodin in complex with the catalytic subunit of protein kinase CK2 | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Niefind, K, Raaf, J, Issinger, O.-G. | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Catalytic Subunit of Human Protein Kinase CK2 Structurally Deviates from Its Maize Homologue in Complex with the Nucleotide Competitive Inhibitor Emodin
J.Mol.Biol., 377, 2008
|
|
3C9A
 
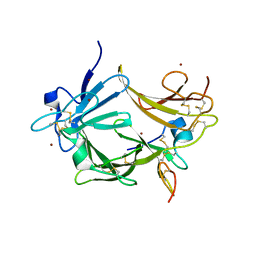 | | High Resolution Crystal Structure of Argos bound to the EGF domain of Spitz | | Descriptor: | BROMIDE ION, Protein giant-lens, Protein spitz | | Authors: | Klein, D.E, Stayrook, S.E, Shi, F, Narayan, K, Lemmon, M.A. | | Deposit date: | 2008-02-15 | | Release date: | 2008-05-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for EGFR ligand sequestration by Argos.
Nature, 453, 2008
|
|
1MV0
 
 | |
3AZE
 
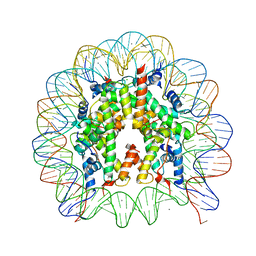 | | Crystal Structure of Human Nucleosome Core Particle Containing H3K64Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
1N21
 
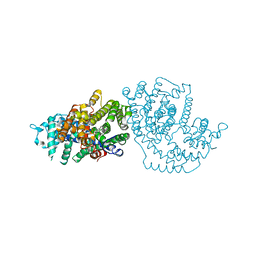 | | (+)-Bornyl Diphosphate Synthase: Cocrystal with Mg and 3-aza-2,3-dihydrogeranyl diphosphate | | Descriptor: | (+)-bornyl diphosphate synthase, 2-[METHYL-(4-METHYL-PENT-3-ENYL)-AMINO]-ETHYL-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Whittington, D.A, Wise, M.L, Urbansky, M, Coates, R.M, Croteau, R.B, Christianson, D.W. | | Deposit date: | 2002-10-21 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Bornyl diphosphate synthase: Structure and strategy for carbocation manipulation by a terpenoid synthase
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N1Z
 
 | | (+)-Bornyl Diphosphate Synthase: Complex with Mg and pyrophosphate | | Descriptor: | (+)-bornyl diphosphate synthase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Whittington, D.A, Wise, M.L, Urbansky, M, Coates, R.M, Croteau, R.B, Christianson, D.W. | | Deposit date: | 2002-10-21 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bornyl diphosphate synthase: Structure and strategy for carbocation manipulation by a terpenoid cyclase
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3U0N
 
 | |
3CA7
 
 | |
3HAV
 
 | | Structure of the streptomycin-ATP-APH(2")-IIa ternary complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aminoglycoside phosphotransferase, MAGNESIUM ION, ... | | Authors: | Young, P.G, Baker, E.N, Vakulenko, S.B, Smith, C.A. | | Deposit date: | 2009-05-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The crystal structures of substrate and nucleotide complexes of Enterococcus faecium aminoglycoside-2''-phosphotransferase-IIa [APH(2'')-IIa] provide insights into substrate selectivity in the APH(2'') subfamily.
J.Bacteriol., 191, 2009
|
|
3U0L
 
 | | Crystal structure of the engineered fluorescent protein mRuby, crystal form 1, pH 4.5 | | Descriptor: | ACETATE ION, mRuby | | Authors: | Akerboom, J, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2011-09-28 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics.
Front Mol Neurosci, 6, 2013
|
|
3U0M
 
 | |
6K50
 
 | |
6QFP
 
 | |
2MN7
 
 | |
6QJL
 
 | |
2M07
 
 | | NMR structure of OmpX in DPC micelles | | Descriptor: | Outer membrane protein X | | Authors: | Hagn, F.X, Etzkorn, M, Raschle, T, Wagner, G, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2012-10-21 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M4J
 
 | | 40-residue beta-amyloid fibril derived from Alzheimer's disease brain | | Descriptor: | Amyloid beta A4 protein | | Authors: | Lu, J, Qiang, W, Meredith, S.C, Yau, W, Schweiters, C.D, Tycko, R. | | Deposit date: | 2013-02-05 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Molecular Structure of beta-Amyloid Fibrils in Alzheimer's Disease Brain Tissue.
Cell(Cambridge,Mass.), 154, 2013
|
|
