6QJY
 
 | |
9FR1
 
 | | Half-closed CODH/ACS in the as-isolated state | | Descriptor: | CO-dehydrogenase, CO-methylating acetyl-CoA synthase, Fe(3)-Ni(1)-S(4) cluster, ... | | Authors: | Ruickoldt, J, Wendler, P, Dobbek, H. | | Deposit date: | 2024-06-18 | | Release date: | 2025-07-02 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Ligand binding to a Ni-Fe cluster orchestrates conformational changes of the CO-dehydrogenase-acetyl-CoA synthase complex.
Nat Catal, 8, 2025
|
|
1G93
 
 | | CRYSTAL STRUCTURE OF THE BOVINE CATALYTIC DOMAIN OF ALPHA-1,3-GALACTOSYLTRANSFERASE IN THE PRESENCE OF UDP-GALACTOSE | | Descriptor: | MANGANESE (II) ION, MERCURY (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Gastinel, L.N, Bignon, C, Misra, A.K, Hindsgaul, O, Shaper, J.H, Joziasse, D.H. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bovine alpha1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases.
EMBO J., 20, 2001
|
|
6GWI
 
 | | The crystal structure of Halomonas elongata amino-transferase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Gourlay, L.J. | | Deposit date: | 2018-06-25 | | Release date: | 2019-05-08 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Widely applicable background depletion step enables transaminase evolution through solid-phase screening.
Chem Sci, 10, 2019
|
|
7UN9
 
 | | SfSTING with c-di-GMP double fiber | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
7UN8
 
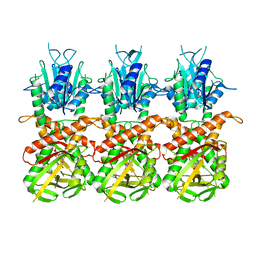 | | SfSTING with c-di-GMP single fiber | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
8KAR
 
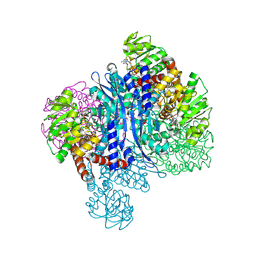 | | Glutamate dehydrogenase-AKG | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, Glutamate dehydrogenase, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural insight into the unique substrate specificity of glutamate dehydrogenase from Saccharolobus solfataricus.
Extremophiles, 29, 2025
|
|
6VQL
 
 | |
5GQK
 
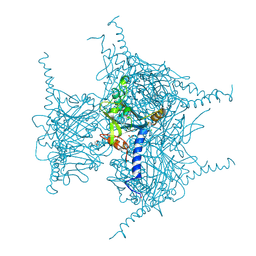 | | Crystal structure of Cypovirus Polyhedra mutant with deletion of Gly192-Ala194 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Polyhedrin | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
5GQN
 
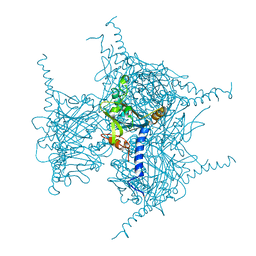 | | Crystal structure of in cellulo Cypovirus Polyhedra mutant with deletion of Gly192-Ala194 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Polyhedrin | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
7VIQ
 
 | | Crystal structure of Au(50EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
6GIO
 
 | |
7W2I
 
 | | Crystal structure of LOG (Rv1205) from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Cytokinin riboside 5'-monophosphate phosphoribohydrolase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shang, L, Zhang, G. | | Deposit date: | 2021-11-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cytokinin-producing enzyme "lonely guy" (LOG) from Mycobacterium tuberculosis.
Biochem.Biophys.Res.Commun., 598, 2022
|
|
5LU6
 
 | | Heptose isomerase mutant - H64Q | | Descriptor: | 1,2-ETHANEDIOL, D-ALTRO-HEPT-2-ULOSE 7-PHOSPHATE, GLYCEROL, ... | | Authors: | Vivoli, M, Harmer, N.J, Pang, J. | | Deposit date: | 2016-09-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A half-site multimeric enzyme achieves its cooperativity without conformational changes.
Sci Rep, 7, 2017
|
|
5LNQ
 
 | |
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7D0O
 
 | | Crystal structure of human HBO1-BRPF2 in apo form | | Descriptor: | 1,2-ETHANEDIOL, BRD1 protein, Histone acetyltransferase KAT7, ... | | Authors: | Li, W, Ding, J. | | Deposit date: | 2020-09-11 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | HBO1 is a versatile histone acyltransferase critical for promoter histone acylations.
Nucleic Acids Res., 49, 2021
|
|
7D0R
 
 | |
5JOE
 
 | | Crystal structure of I81 from titin | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Titin | | Authors: | Fleming, J, Zhou, T, Bogomolovas, J, Labeit, S, Mayans, O. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CARP interacts with titin at a unique helical N2A sequence and at the domain Ig81 to form a structured complex.
Febs Lett., 590, 2016
|
|
5J2W
 
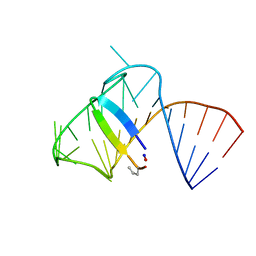 | | Intermediate state lying on the pathway of release of Tat from HIV-1 TAR. | | Descriptor: | Apical region (29mer) of the HIV-1 TAR RNA element, Cyclic peptide mimetic of HIV-1 Tat | | Authors: | Borkar, A.N, Bardaro Jr, M.F, Varani, G, Vendruscolo, M. | | Deposit date: | 2016-03-30 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of a low-population binding intermediate in protein-RNA recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J46
 
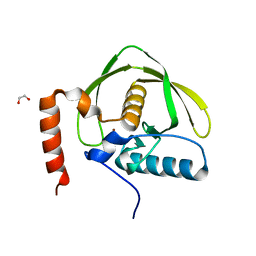 | |
6DDQ
 
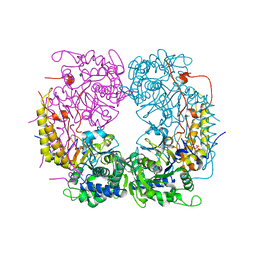 | | Crystal structure of the double mutant (R39Q/D52N) of the full-length NT5C2 in the basal state | | Descriptor: | 1,2-ETHANEDIOL, Cytosolic purine 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
1P3X
 
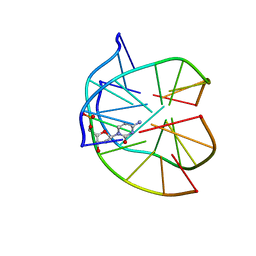 | | INTRAMOLECULAR DNA TRIPLEX WITH 1-PROPYNYL DEOXYURIDINE IN THE THIRD STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*(PDU)P*CP*(PDU)P*(DCM)P*(PDU)P*CP*(PDU)P*(PDU))-3'), DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*CP*T)-3') | | Authors: | Phipps, A.K, Tarkoy, M, Schultze, P, Feigon, J. | | Deposit date: | 1998-02-05 | | Release date: | 1998-05-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an intramolecular DNA triplex containing 5-(1-propynyl)-2'-deoxyuridine residues in the third strand.
Biochemistry, 37, 1998
|
|
1Y66
 
 | | Dioxane contributes to the altered conformation and oligomerization state of a designed engrailed homeodomain variant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACETIC ACID, CADMIUM ION, ... | | Authors: | Hom, G.K, Lassila, J.K, Thomas, L.M, Mayo, S.L. | | Deposit date: | 2004-12-03 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dioxane contributes to the altered conformation and oligomerization state of a designed engrailed homeodomain variant.
Protein Sci., 14, 2005
|
|
8RH6
 
 | | Crystal structure of HLA-A*11:01 in complex with SVLNDILSRL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
