1T51
 
 | | Antibiotic Activity and Structural Analysis of a Scorpion-derived Antimicrobial peptide IsCT and Its Analogs | | Descriptor: | Cytotoxic linear peptide IsCT | | Authors: | Lee, K, Shin, S.Y, Kim, K, Lim, S.S, Hahm, K.S, Kim, Y. | | Deposit date: | 2004-05-01 | | Release date: | 2004-10-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs
Biochem.Biophys.Res.Commun., 323, 2004
|
|
1HKQ
 
 | | PPS10 plasmid DNA replication initiator protein RepA. Replication inactive, dimeric N-terminal domain. | | Descriptor: | BENZOIC ACID, MERCURY (II) ION, PHOSPHATE ION, ... | | Authors: | Giraldo, R, Fernandez-Tornero, C, Evans, P.R, Diaz-Orejas, R, Romero, A. | | Deposit date: | 2003-03-11 | | Release date: | 2003-05-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A Conformational Switch between Transcriptional Repression and Replication Initiation in Repa Dimerization Domain
Nat.Struct.Biol., 10, 2003
|
|
1T5S
 
 | | Structure of the (SR)Ca2+-ATPase Ca2-E1-AMPPCP form | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Sorensen, T.L.-M, Moller, J.V, Nissen, P. | | Deposit date: | 2004-05-05 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phosphoryl transfer and calcium ion occlusion in the calcium pump.
Science, 304, 2004
|
|
1T86
 
 | | Crystal Structure of the Ferrous Cytochrome P450cam Mutant (L358P/C334A) | | Descriptor: | CAMPHOR, Cytochrome P450-cam, POTASSIUM ION, ... | | Authors: | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | Deposit date: | 2004-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
1TAS
 
 | |
1TE0
 
 | |
1IIL
 
 | | CRYSTAL STRUCTURE OF PRO253ARG APERT MUTANT FGF RECEPTOR 2 (FGFR2) IN COMPLEX WITH FGF2 | | Descriptor: | FIBROBLAST GROWTH FACTOR RECEPTOR 2, HEPARIN-BINDING GROWTH FACTOR 2 | | Authors: | Ibrahimi, O.A, Eliseenkova, A.V, Plotnikov, A.N, Ornitz, D.M, Mohammadi, M. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for fibroblast growth factor receptor 2 activation in Apert syndrome.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IUK
 
 | | The structure of native ID.343 from Thermus thermophilus | | Descriptor: | hypothetical protein TT1466 | | Authors: | Wada, T, Shirouzu, M, Park, S.-Y, Tame, J.R, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-05 | | Release date: | 2003-07-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a conserved CoA-binding protein synthesized by a cell-free system.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1IUU
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE AT PH 9.4 | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
1SZW
 
 | |
1J2C
 
 | | Crystal structure of rat heme oxygenase-1 in complex with biliverdin IXalpha-iron cluster | | Descriptor: | BILIVERDINE IX ALPHA, FE (III) ION, Heme Oxygenase-1 | | Authors: | Sugishima, M, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2002-12-29 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Rat Heme Oxygenase-1 in Complex with Biliverdin-Iron Chelate: CONFORMATIONAL CHANGE OF THE DISTAL HELIX DURING THE HEME CLEAVAGE REACTION.
J.Biol.Chem., 278, 2003
|
|
1T5N
 
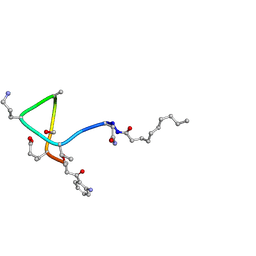 | | Structural transitions as determinants of calcium-dependent antibiotic daptomycin | | Descriptor: | DAPTOMYCIN, DECANOIC ACID | | Authors: | Jung, D, Rozek, A, Okon, M, Hancock, R.E. | | Deposit date: | 2004-05-04 | | Release date: | 2004-08-31 | | Last modified: | 2019-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Transitions as Determinants of the Action of the Calcium-Dependent Antibiotic Daptomycin.
Chem.Biol., 11, 2004
|
|
1J5D
 
 | | SOLUTION STRUCTURE OF OXIDIZED PARAMAGNETIC CU(II) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803-MINIMIZED AVERAGE STRUCTURE | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Shumilin, S, Vila, A.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-04-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The first solution structure of a paramagnetic copper(II) protein: the case of oxidized plastocyanin from the cyanobacterium Synechocystis PCC6803.
J.Am.Chem.Soc., 123, 2001
|
|
1T5T
 
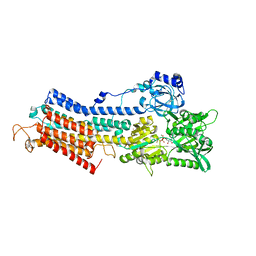 | | Structure of the (SR)Ca2+-ATPase Ca2-E1-ADP:AlF4- form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Sorensen, T.L.-M, Moller, J.V, Nissen, P. | | Deposit date: | 2004-05-05 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Phosphoryl transfer and calcium ion occlusion in the calcium pump.
Science, 304, 2004
|
|
1T1Q
 
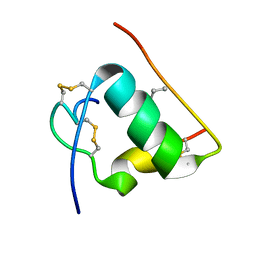 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT HIS-B10-ASP, VAL-B12-ABA, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin, insulin | | Authors: | Huang, K, Xu, B, Hu, S.Q, Chu, Y.C, Hua, Q.X, Whittaker, J, Nakagawa, S.H, De Meyts, P, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-10 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | How Insulin Binds: the B-Chain alpha-Helix Contacts the L1 beta-Helix of the Insulin Receptor.
J.Mol.Biol., 341, 2004
|
|
1T3K
 
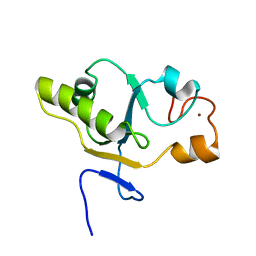 | | NMR structure of a CDC25-like dual-specificity tyrosine phosphatase of Arabidopsis thaliana | | Descriptor: | Dual-specificity tyrosine phosphatase, ZINC ION | | Authors: | Landrieu, I, da Costa, M, De Veylder, L, Dewitte, F, Vandepoele, K, Hassan, S, Wieruszeski, J.M, Faure, J.D, Inze, D, Lippens, G. | | Deposit date: | 2004-04-27 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A small CDC25 dual-specificity tyrosine-phosphatase isoform in Arabidopsis thaliana.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1T87
 
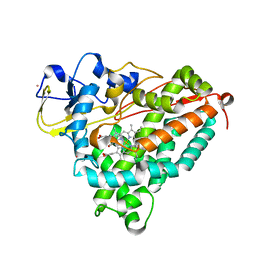 | | Crystal Structure of the Ferrous CO-bound Cytochrome P450cam (C334A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CARBON MONOXIDE, ... | | Authors: | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | Deposit date: | 2004-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
1T52
 
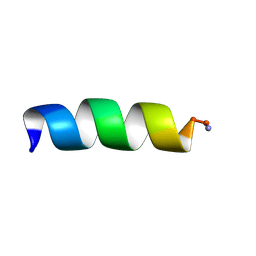 | | Antibiotic Activity and Structural Analysis of a Scorpion-derived Antimicrobial peptide IsCT and Its Analogs | | Descriptor: | Cytotoxic linear peptide IsCT | | Authors: | Lee, K, Shin, S.Y, Kim, K, Lim, S.S, Hahm, K.S, Kim, Y. | | Deposit date: | 2004-05-01 | | Release date: | 2004-10-19 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs
Biochem.Biophys.Res.Commun., 323, 2004
|
|
1JEQ
 
 | |
1T88
 
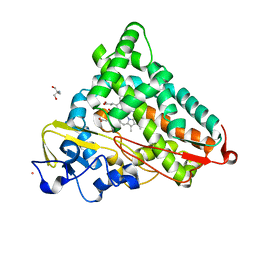 | | Crystal Structure of the Ferrous Cytochrome P450cam (C334A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Cytochrome P450-cam, ... | | Authors: | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | Deposit date: | 2004-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
1W7U
 
 | |
1W29
 
 | | Lumazine Synthase from Mycobacterium tuberculosis bound to 3-(1,3,7- trihydro-9-D-ribityl-2,6,8-purinetrione-7-yl)butane 1-phosphate | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 4-{2,6,8-TRIOXO-9-[(2R,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL]-1,2,3,6,8,9-HEXAHYDRO-7H-PURIN-7-YL}BUTYL DIHYDROGEN PHOSPHATE, 4-{2,6,8-TRIOXO-9-[(2S,3R,4R)-2,3,4,5-TETRAHYDROXYPENTYL]-1,2,3,6,8,9-HEXAHYDRO-7H-PURIN-7-YL}BUTYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Morgunova, E, Meining, W, Illarionov, B, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2004-07-01 | | Release date: | 2005-03-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Lumazine Synthase from Mycobacterium Tuberculosis as a Target for Rational Drug Design: Binding Mode of a New Class of Purinetrione Inhibitors(,)
Biochemistry, 44, 2005
|
|
1W93
 
 | | Crystal Structure of Biotin Carboxylase Domain of Acetyl-Coenzyme A Carboxylase from Saccharomyces cerevisiae | | Descriptor: | ACETYL-COENZYME A CARBOXYLASE | | Authors: | Shen, Y, Volrath, S.L, Weatherly, S.C, Elich, T.D, Tong, L. | | Deposit date: | 2004-10-05 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Mechanism for the Potent Inhibition of Eukaryotic Acetyl-Coenzyme a Carboxylase by Soraphen A, a Macrocyclic Polyketide Natural Product
Mol.Cell, 16, 2004
|
|
1K0L
 
 | | Pseudomonas aeruginosa phbh R220Q free of p-OHB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, SULFATE ION, ... | | Authors: | Wang, J, Ortiz-Maldonado, M, Entsch, B, Ballou, D, Gatti, D.L. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K2H
 
 | | Three-dimensional Solution Structure of apo-S100A1. | | Descriptor: | S-100 protein, alpha chain | | Authors: | Rustandi, R.R, Baldisseri, D.M, Inman, K.G, Nizner, P, Hamilton, S.M, Landar, A, Landar, A, Zimmer, D.B, Weber, D.J. | | Deposit date: | 2001-09-27 | | Release date: | 2002-02-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium-signaling protein apo-S100A1 as determined by NMR.
Biochemistry, 41, 2002
|
|
