6HT6
 
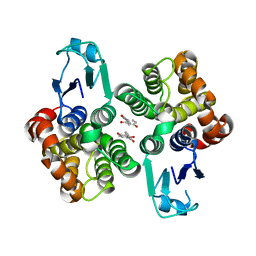 | | Crystal structure of glutathione transferase Omega 2S from Trametes versicolor in complex with 2,4-dihydroxybenzophenone | | Descriptor: | Glutathione S-transferase omega 2S, MAGNESIUM ION, [2,4-bis(oxidanyl)phenyl]-phenyl-methanone | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.671 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
6ZLR
 
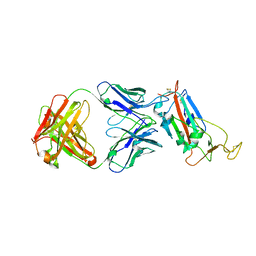 | |
6H7X
 
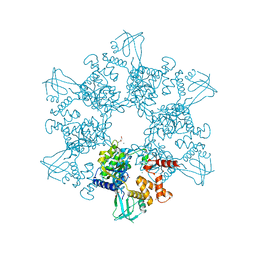 | | First X-ray structure of full-length human RuvB-Like 2. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Silva, S, Brito, J.A, Matias, P, Bandeiras, T. | | Deposit date: | 2018-07-31 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | X-ray structure of full-length human RuvB-Like 2 - mechanistic insights into coupling between ATP binding and mechanical action.
Sci Rep, 8, 2018
|
|
6QGX
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody F7 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoF7, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
8TJI
 
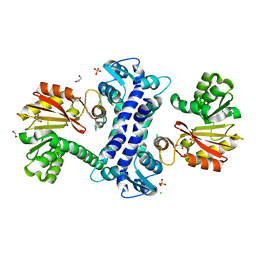 | | SAM-dependent methyltransferase RedM, apo | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, RedM, ... | | Authors: | Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2023-07-22 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure of methyltransferase RedM that forms the dimethylpyrrolinium of the bisindole reductasporine.
J.Biol.Chem., 300, 2023
|
|
7TDL
 
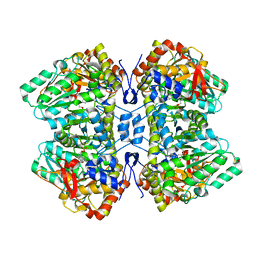 | | M379A mutant tyrosine phenol-lyase complexed with 3-bromo-DL-phenylalanine | | Descriptor: | (4Z)-4-({[(1E)-2-(3-bromophenyl)-1-carboxyethylidene]azaniumyl}methylidene)-2-methyl-5-[(phosphonooxy)methyl]-1,4-dihydropyridin-3-olate, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | M379A Mutant Tyrosine Phenol-lyase from Citrobacter freundii Has Altered Conformational Dynamics.
Chembiochem, 23, 2022
|
|
6Y4A
 
 | |
6G4B
 
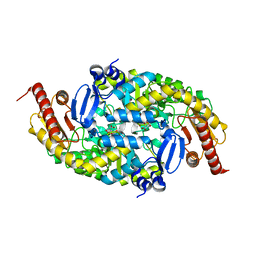 | |
7T7O
 
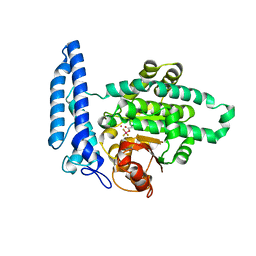 | |
6YD1
 
 | | SaFtsZ-DFMBA | | Descriptor: | 2,6-difluoro-3-methoxybenzamide, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
6QH9
 
 | | Crystal Structure of Human Kallikrein 6 in complex with GSK3239861A | | Descriptor: | (3~{R})-~{N}-(4-carbamimidoylphenyl)-2-oxidanylidene-piperidine-3-carboxamide, (3~{S})-~{N}-(4-carbamimidoylphenyl)-2-oxidanylidene-piperidine-3-carboxamide, GLYCEROL, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-06 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Kallikrein 5 inhibitors identified through structure based drug design in search for a treatment for Netherton Syndrome.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6H8A
 
 | | MamM CTD D249N - Cadmium form | | Descriptor: | BETA-MERCAPTOETHANOL, CADMIUM ION, Magnetosome protein MamM, ... | | Authors: | Barber-Zucker, S, Zarivach, R. | | Deposit date: | 2018-08-02 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MamM CTD D249N - Cadmium form
To Be Published
|
|
7T7K
 
 | | Structure of SPAC806.04c protein from fission yeast bound to Co2+ | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Damage-control phosphatase SPAC806.04c, ... | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
6YD6
 
 | | SaFtsZ-UCM152 (comp.20) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylpyrrolidin-2-one, 2,6-bis(fluoranyl)-3-[[3-(trifluoromethyl)phenyl]methoxy]benzamide, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
6G4N
 
 | | Torpedo californica acetylcholinesterase bound to uncharged hybrid reactivator 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[4-[(7-chloranyl-1,2,3,4-tetrahydroacridin-9-yl)amino]butyl]-2-[(oxidanylamino)methyl]pyridin-3-ol, Acetylcholinesterase, ... | | Authors: | Santoni, G, De la Mora, E, de Souza, J, Silman, I, Sussman, J, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
8TGB
 
 | | Crystal structure of root lateral formation protein (RLF) b5-domain from Oryza sativa | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, root lateral formation protein (RLF) | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Benson, D.R. | | Deposit date: | 2023-07-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The N-terminal intrinsically disordered region of Ncb5or docks with the cytochrome b 5 core to form a helical motif that is of ancient origin.
Proteins, 92, 2024
|
|
6ZR7
 
 | | X-ray structure of human Dscam Ig7-Ig9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Down syndrome cell adhesion molecule, ... | | Authors: | Kozak, S, Bento, I, Meijers, R. | | Deposit date: | 2020-07-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Homogeneously N-glycosylated proteins derived from the GlycoDelete HEK293 cell line enable diffraction-quality crystallogenesis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6YD5
 
 | | SaFtsZ-UCM151 (comp. 18) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylpyrrolidin-2-one, 3-[(3-chlorophenyl)methoxy]-2,6-bis(fluoranyl)benzamide, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
6QHJ
 
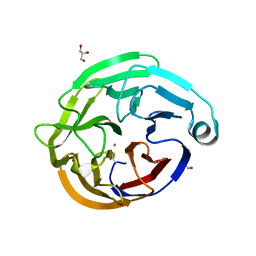 | | High-resolution crystal structure of calcium- and sodium-bound mouse Olfactomedin-1 beta-propeller domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Pronker, M.F, van den Hoek, H.G, Janssen, B.J.C. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and structural characterisation of olfactomedin-1 variants as tools for functional studies.
BMC Mol Cell Biol, 20, 2019
|
|
6H8G
 
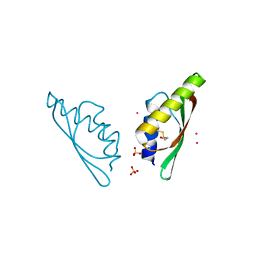 | | MamM CTD H264E - Cadmium form 1 | | Descriptor: | BETA-MERCAPTOETHANOL, CADMIUM ION, Magnetosome protein MamM, ... | | Authors: | Barber-Zucker, S, Zarivach, R. | | Deposit date: | 2018-08-02 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | MamM CTD H264E - Cadmium form 1
To Be Published
|
|
6FUR
 
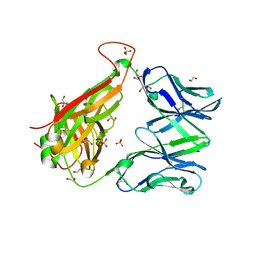 | |
7ZJ3
 
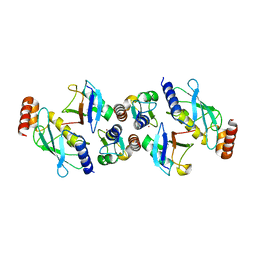 | | Structure of TRIM2 RING domain in complex with UBE2D1~Ub conjugate | | Descriptor: | Polyubiquitin-C, Tripartite motif-containing protein 2, Ubiquitin-conjugating enzyme E2 D1, ... | | Authors: | Esposito, D, Garza-Garcia, A, Dudley-Fraser, J, Rittinger, K. | | Deposit date: | 2022-04-08 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Divergent self-association properties of paralogous proteins TRIM2 and TRIM3 regulate their E3 ligase activity.
Nat Commun, 13, 2022
|
|
8TJK
 
 | | SAM-dependent methyltransferase RedM bound to SAH | | Descriptor: | CHLORIDE ION, RedM, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2023-07-22 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of methyltransferase RedM that forms the dimethylpyrrolinium of the bisindole reductasporine.
J.Biol.Chem., 300, 2023
|
|
8TJJ
 
 | | SAM-dependent methyltransferase RedM bound to SAM | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, RedM, ... | | Authors: | Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2023-07-22 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of methyltransferase RedM that forms the dimethylpyrrolinium of the bisindole reductasporine.
J.Biol.Chem., 300, 2023
|
|
7SPD
 
 | |
