7LWC
 
 | | Goat beta-lactoglobulin mutant Q59A | | Descriptor: | Beta-lactoglobulin | | Authors: | Munoz, D.A, Dobson, R.C.J. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineering food proteins for improved digestibility and lower allergenicity: a case study using caprine beta-lactoglobulin
To Be Published
|
|
7QWK
 
 | | GCN2 (EIF2ALPHA KINASE 4, E2AK4) IN COMPLEX WITH COMPOUND 2 | | Descriptor: | (2~{S})-~{N}-[(1~{S})-1-[4-[(6-pyridin-4-ylquinazolin-2-yl)amino]phenyl]ethyl]piperidine-2-carboxamide, DIMETHYL SULFOXIDE, eIF-2-alpha kinase GCN2 | | Authors: | Maia de Oliveira, T. | | Deposit date: | 2022-01-25 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of human GCN2 reveals a parallel, back-to-back kinase dimer with a plastic DFG activation loop motif.
Biochem.J., 477, 2020
|
|
7QS2
 
 | |
6BID
 
 | | 1.15 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | Descriptor: | 3C-like protease, benzyl [(8S,11S,14S)-11-(cyclohexylmethyl)-8-(hydroxymethyl)-5,10,13-trioxo-1,4,9,12,17,18-hexaazabicyclo[14.2.1]nonadeca-16(19),17-dien-14-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
6BIM
 
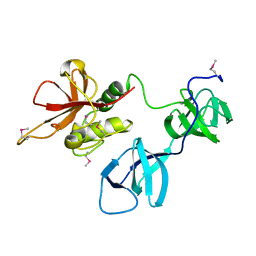 | |
6BJH
 
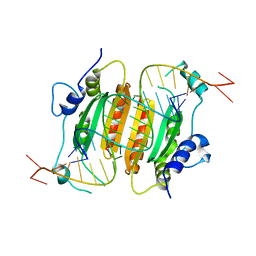 | | CIRV p19 mutant T111S in complex with siRNA | | Descriptor: | RNA (5'-R(P*CP*GP*UP*AP*CP*GP*CP*GP*GP*AP*AP*UP*AP*CP*UP*UP*CP*GP*AP*UP*U)-3'), RNA (5'-R(P*UP*CP*GP*AP*AP*GP*UP*AP*UP*UP*CP*CP*GP*CP*GP*UP*AP*CP*GP*UP*U)-3'), RNA silencing suppressor p19 | | Authors: | Foss, D.V, Schirle, N.T, MacRae, I.J, Pezacki, J.P. | | Deposit date: | 2017-11-06 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural insights into interactions between viral suppressor of RNA silencing protein p19 mutants and small RNAs.
Febs Open Bio, 9, 2019
|
|
7QG3
 
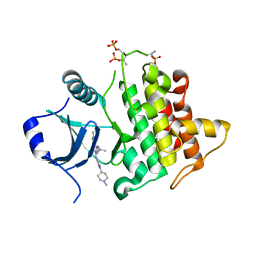 | | IRAK4 in complex with inhibitor | | Descriptor: | 6-[(2~{S})-2-fluoranylpropyl]-4-[(1-methylcyclopropyl)amino]-2-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]pyrido[4,3-d]pyrimidin-5-one, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
7L5S
 
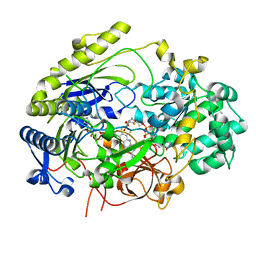 | | Crystal Structure of Haemophilus influenzae MtsZ at pH 5.5 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, MOLYBDENUM ATOM, OXYGEN ATOM, ... | | Authors: | Struwe, M.A, Luo, Z, Kappler, U, Kobe, B. | | Deposit date: | 2020-12-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Active site architecture reveals coordination sphere flexibility and specificity determinants in a group of closely related molybdoenzymes.
J.Biol.Chem., 296, 2021
|
|
7YVV
 
 | | AcmP1, R-4-hydroxymandelate synthase | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, CHLORIDE ION, FE (III) ION, ... | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2022-08-19 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of AcmP1 as the native R-4-hydroxymandelate synthase from biosynthetic pathway of Amycolamycins
To Be Published
|
|
7QRX
 
 | |
6ZTH
 
 | |
6IIW
 
 | | Crystal structure of human UHRF1 PHD finger in complex with PAF15 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase UHRF1, PCNA-associated factor, ... | | Authors: | Arita, K, Kori, S. | | Deposit date: | 2018-10-07 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Two distinct modes of DNMT1 recruitment ensure stable maintenance DNA methylation.
Nat Commun, 11, 2020
|
|
6BP0
 
 | | Crystal Structure of the Human vaccinia-related kinase 1 bound to (R)-2-phenylaminopteridinone inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7,8-trimethyl-7,8-dihydropteridin-6(5H)-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase 1 bound to (R)-2-phenylaminopteridinone inhibitor
To Be Published
|
|
7YVY
 
 | |
7QS5
 
 | |
6BKU
 
 | | Crystal Structure of the Human CAMKK2B bound to GSK650394 | | Descriptor: | 2-cyclopentyl-4-(5-phenyl-1H-pyrrolo[2,3-b]pyridin-3-yl)benzoic acid, Calcium/calmodulin-dependent protein kinase kinase 2, FORMIC ACID | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Ramos, P.Z, Drewry, D, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-09 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human CAMKK2B bound to GSK650394
To Be Published
|
|
7QS3
 
 | |
7YUJ
 
 | | Crystal structure of HOIL-1L(365-510) | | Descriptor: | DI(HYDROXYETHYL)ETHER, RanBP-type and C3HC4-type zinc finger-containing protein 1, ZINC ION | | Authors: | Xiao, L, Pan, L. | | Deposit date: | 2022-08-17 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.865 Å) | | Cite: | Mechanistic insights into the enzymatic activity of E3 ligase HOIL-1L and its regulation by the linear ubiquitin chain binding.
Sci Adv, 9, 2023
|
|
7QT8
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound ABT-957 | | Descriptor: | (2~{R})-5-oxidanylidene-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]-1-(phenylmethyl)pyrrolidine-2-carboxamide, 3C-like proteinase | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7YUI
 
 | |
6BLH
 
 | | RSV G central conserved region bound to Fab CB017.5 | | Descriptor: | 1,2-ETHANEDIOL, Fab CB017.5 heavy chain, Fab CB017.5 light chain, ... | | Authors: | Jones, H.G, McLellan, J.S, Langedijk, J.P. | | Deposit date: | 2017-11-10 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for recognition of the central conserved region of RSV G by neutralizing human antibodies.
PLoS Pathog., 14, 2018
|
|
7QT6
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z1367324110 | | Descriptor: | 1-methyl-3,4-dihydro-2~{H}-quinoline-7-sulfonamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QT5
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z31792168 | | Descriptor: | 2-cyclohexyl-~{N}-pyridin-3-yl-ethanamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8R3E
 
 | | Huntingtin, 1-17, MBP-N | | Descriptor: | Maltodextrin-binding protein, ZINC ION | | Authors: | Steinbacher, S, Toledo-Sherman, L, Dominguez, C. | | Deposit date: | 2023-11-08 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Post-translational modifications in the first 17 amino acids of huntingtin influence self-association and interaction with membranes
To Be Published
|
|
7YRS
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MOPS | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
