8CCR
 
 | |
5EZE
 
 | |
5EZ9
 
 | |
5F2Y
 
 | |
5EZ8
 
 | |
5EZA
 
 | |
5EZC
 
 | |
6LLQ
 
 | | Solution NMR structure of de novo Rossmann2x2 fold with most of the core mutated to valine, R2x2_VAL88 | | Descriptor: | VAL88 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Koga, R, Yamamoto, M, Kosugi, T, Koga, N. | | Deposit date: | 2019-12-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Robust folding of a de novo designed ideal protein even with most of the core mutated to valine.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5EON
 
 | |
6W70
 
 | | Crystal Structure of apixaban-bound ABLE | | Descriptor: | 1-(4-METHOXYPHENYL)-7-OXO-6-[4-(2-OXOPIPERIDIN-1-YL)PHENYL]-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDINE-3-CARBOXAMIDE, ACETATE ION, De novo designed ABLE, ... | | Authors: | Polizzi, N.F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | A defined structural unit enables de novo design of small-molecule-binding proteins.
Science, 369, 2020
|
|
6W6X
 
 | | Crystal Structure of ABLE Apo-protein | | Descriptor: | ACETATE ION, De novo designed ABLE protein, SULFATE ION | | Authors: | Polizzi, N.F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | A defined structural unit enables de novo design of small-molecule-binding proteins.
Science, 369, 2020
|
|
5EHB
 
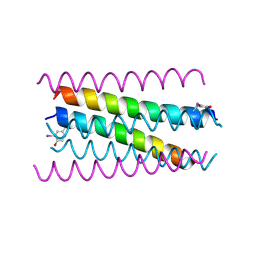 | | A de novo designed hexameric coiled-coil peptide with iodotyrosine | | Descriptor: | pHiosYI | | Authors: | Lizatovic, R, Aurelius, O, Stenstrom, O, Drakenberg, T, Akke, M, Logan, D.T, Andre, I. | | Deposit date: | 2015-10-28 | | Release date: | 2016-06-15 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | A De Novo Designed Coiled-Coil Peptide with a Reversible pH-Induced Oligomerization Switch.
Structure, 24, 2016
|
|
6Z0L
 
 | | Het-N2 - De novo designed three-helix heterodimer with Cysteine at the N2 position of the alpha-helix | | Descriptor: | CADMIUM ION, Cys-N2 Strand, Positive Strand, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | Deposit date: | 2020-05-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
6Z0M
 
 | | Het-Ncap - De novo designed three-helix heterodimer with Cysteine at the Ncap position of the alpha-helix | | Descriptor: | Cys-Ncap strand, Positive Strand, SULFATE ION, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | Deposit date: | 2020-05-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
5JI4
 
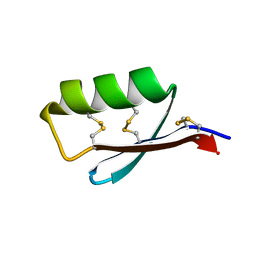 | | Solution structure of the de novo mini protein gEEHE_02 | | Descriptor: | W37 | | Authors: | Buchko, G.W, Bahl, C.D, Pulavarti, S.V, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
7BEY
 
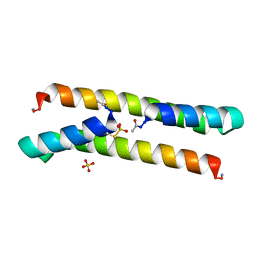 | | Het-N2-SO3- - De novo designed three-helix heterodimer with Cysteine S-sulfate at the N2 position of the alpha-helix | | Descriptor: | 'Cys-N2-SO3-' Strand, 'Positive' Strand, SULFATE ION | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
7BIM
 
 | | A de novo designed nonameric coiled coil, CC-Type2-(GgLaId)4 | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Nonameric de novo coiled coil CC-Type2-(GgLaId)4 | | Authors: | Shelley, K.L, Rhys, G.G, Dawson, W.M, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2021-01-12 | | Release date: | 2021-04-21 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Coiled coils 9-to-5: rational de novo design of alpha-helical barrels with tunable oligomeric states.
Chem Sci, 12, 2021
|
|
8UGC
 
 | |
5JHI
 
 | | Solution structure of the de novo mini protein gEHE_06 | | Descriptor: | W35 | | Authors: | Buchko, G.W, Bahl, C.D, Gilmore, J.M, Pulavarti, S.V, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
7OSU
 
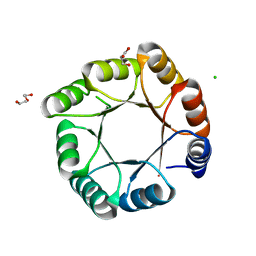 | | sTIM11noCys-SB, a de novo designed TIM barrel with a salt-bridge cluster (crystal form 1) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Romero-Romero, S, Kordes, S, Hocker, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A newly introduced salt bridge cluster improves structural and biophysical properties of de novo TIM barrels.
Protein Sci., 31, 2022
|
|
7OT7
 
 | |
3VJF
 
 | | Crystal structure of de novo 4-helix bundle protein WA20 | | Descriptor: | POTASSIUM ION, WA20 | | Authors: | Arai, R, Kimura, A, Kobayashi, N, Matsuo, K, Sato, T, Wang, A.F, Platt, J.M, Bradley, L.H, Hecht, M.H. | | Deposit date: | 2011-10-18 | | Release date: | 2012-03-28 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Domain-swapped dimeric structure of a stable and functional de novo four-helix bundle protein, WA20
J.Phys.Chem.B, 116, 2012
|
|
3P6I
 
 | |
7AH0
 
 | | Crystal structure of the de novo designed two-heme binding protein, 4D2 | | Descriptor: | 4D2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hutchins, G.H, Parnell, A.E, Anderson, J.L.R. | | Deposit date: | 2020-09-23 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An expandable, modular de novo protein platform for precision redox engineering.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3RA3
 
 | | Crystal structure of a section of a de novo design gigaDalton protein fibre | | Descriptor: | SODIUM ION, p1c, p2f | | Authors: | Zaccai, N.R, Sharp, T.H, Bruning, M, Woolfson, D.N, Brady, R.L. | | Deposit date: | 2011-03-27 | | Release date: | 2012-08-08 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Cryo-transmission electron microscopy structure of a gigadalton peptide fiber of de novo design
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
