1HD0
 
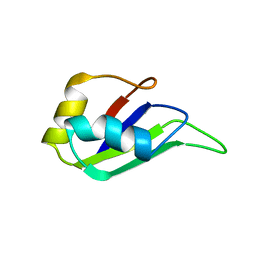 | | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0 (HNRNP D0 RBD1), NMR | | Descriptor: | PROTEIN (HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0) | | Authors: | Nagata, T, Kurihara, Y, Matsuda, G, Saeki, J, Kohno, T, Yanagida, Y, Ishikawa, F, Uesugi, S, Katahira, M. | | Deposit date: | 1999-05-18 | | Release date: | 2000-05-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions with RNA of the N-terminal UUAG-specific RNA-binding domain of hnRNP D0.
J.Mol.Biol., 287, 1999
|
|
1HA1
 
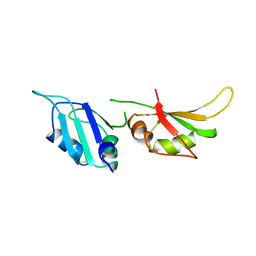 | | HNRNP A1 (RBD1,2) FROM HOMO SAPIENS | | Descriptor: | HNRNP A1 | | Authors: | Shamoo, Y, Krueger, U, Rice, L, Williams, K.R, Steitz, T.A. | | Deposit date: | 1996-10-30 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 A resolution.
Nat.Struct.Biol., 4, 1997
|
|
1HD1
 
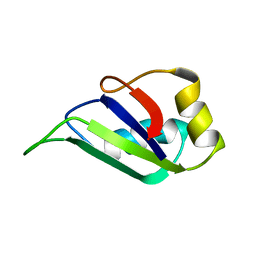 | | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0 (HNRNP D0 RBD1), NMR | | Descriptor: | PROTEIN (HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0) | | Authors: | Nagata, T, Kurihara, Y, Matsuda, G, Saeki, J, Kohno, T, Yanagida, Y, Ishikawa, F, Uesugi, S, Katahira, M. | | Deposit date: | 1999-05-18 | | Release date: | 2000-05-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions with RNA of the N-terminal UUAG-specific RNA-binding domain of hnRNP D0.
J.Mol.Biol., 287, 1999
|
|
6Z29
 
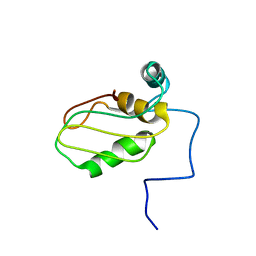 | | Structure of eIF4G1 (37-49) - PUB1 RRM3 chimera in solution | | Descriptor: | Eukaryotic initiation factor 4F subunit p150,Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1 | | Authors: | Chaves-Arquero, B, Martinez-Lumbreras, S, Perez-Canadillas, J.M. | | Deposit date: | 2020-05-15 | | Release date: | 2021-05-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | eIF4G1 N-terminal intrinsically disordered domain is a multi-docking station for RNA, Pab1, Pub1, and self-assembly.
Front Mol Biosci, 9, 2022
|
|
8ILZ
 
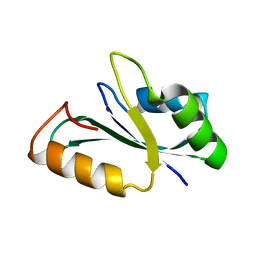 | | Crystal structure of the RRM domain of human SETD1B | | Descriptor: | Histone-lysine N-methyltransferase SETD1B | | Authors: | Bao, S, Xu, C. | | Deposit date: | 2023-03-05 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Molecular insight into the SETD1A/B N-terminal region and its interaction with WDR82.
Biochem.Biophys.Res.Commun., 658, 2023
|
|
8ILY
 
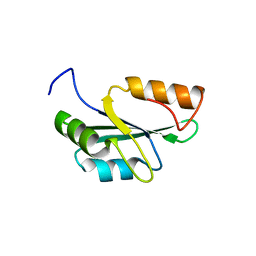 | | Crystal structure of the RRM domain of human SETD1A | | Descriptor: | SET domain containing 1A, histone lysine methyltransferase | | Authors: | Bao, S, Xu, C. | | Deposit date: | 2023-03-05 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular insight into the SETD1A/B N-terminal region and its interaction with WDR82.
Biochem.Biophys.Res.Commun., 658, 2023
|
|
7WM3
 
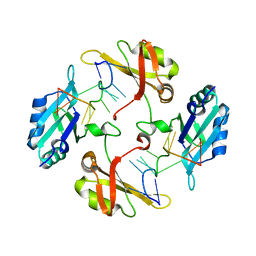 | | hnRNP A2/B1 RRMs in complex with single-stranded DNA | | Descriptor: | DNA (5'-D(P*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Abula, A, Liu, Y, Guo, H, Li, T, Ji, X. | | Deposit date: | 2022-01-14 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Insight Into hnRNP A2/B1 Homodimerization and DNA Recognition.
J.Mol.Biol., 435, 2022
|
|
7VSJ
 
 | |
7VRL
 
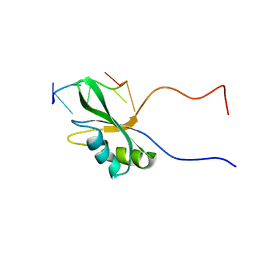 | | Solution structure of Rbfox RRM bound to a non-cognate RNA | | Descriptor: | RNA (5'-R(*UP*GP*CP*AP*UP*AP*U)-3'), RNA binding protein fox-1 homolog 1 | | Authors: | Yang, F, Varani, G. | | Deposit date: | 2021-10-23 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two distinct binding modes provide the RNA-binding protein RbFox with extraordinary sequence specificity.
Nat Commun, 14, 2023
|
|
7WEZ
 
 | |
7YEW
 
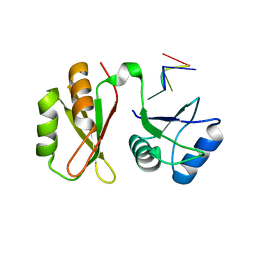 | | Structure of MmIGF2BP3 in complex with 7-mer RNA | | Descriptor: | Insulin-like growth factor 2 mRNA-binding protein 3, RNA (5'-R(P*CP*AP*CP*AP*CP*AP*C)-3') | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of MmIGF2BP3 in complex with 7-mer RNA
To Be Published
|
|
7YEX
 
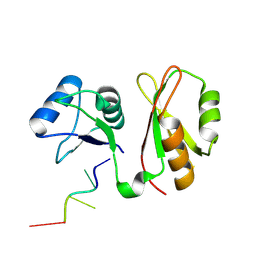 | | Structure of MmIGF2BP3 in complex with 8-mer RNA | | Descriptor: | Insulin-like growth factor 2 mRNA-binding protein 3, RNA (5'-R(P*CP*AP*CP*AP*CP*AP*CP*A)-3') | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of MmIGF2BP3 in complex with 8-mer RNA
To Be Published
|
|
7XX9
 
 | |
7XX8
 
 | |
7ZEV
 
 | | Free form of extended Cyp33-RRM | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7ZEW
 
 | | Complex Cyp33-RRM : AAUAAA RNA | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E, RNA (5'-R(*AP*AP*UP*AP*AP*A)-3') | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7ZEX
 
 | | Complex Cyp33-RRMdelta alpha : UAAUGUCG RNA | | Descriptor: | Isoform 3 of Peptidyl-prolyl cis-trans isomerase E, RNA (5'-R(*UP*AP*AP*UP*GP*UP*CP*G)-3') | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7Q4L
 
 | |
7Q33
 
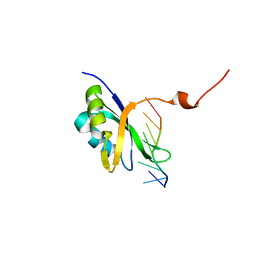 | |
7QZP
 
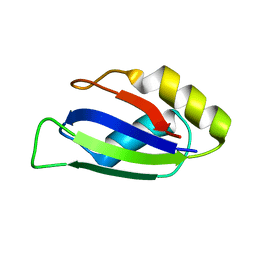 | | Identification and characterization of an RRM-containing, ELAV-like, RNA binding protein in Acinetobacter Baumannii | | Descriptor: | Hypothetical RNA binding protein from Acinetobacter baumannii | | Authors: | Ciani, C, Perez-Rafols, A, Bonomo, I, Micaelli, M, Esposito, A, Zucal, C, Belli, R, D'Agostino, V.G, Bianconi, I, Calderone, V, Cerofolini, L, Fragai, M, Provenzani, A. | | Deposit date: | 2022-01-31 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification and Characterization of an RRM-Containing, RNA Binding Protein in Acinetobacter baumannii .
Biomolecules, 12, 2022
|
|
7Q8A
 
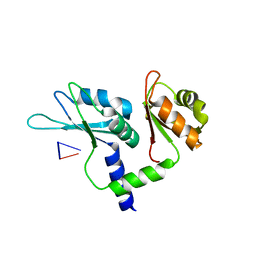 | | Crystal structure of tandem domain RRM1-2 of FUBP-interacting repressor (FIR) bound to FUSE ssDNA fragment | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*GP*T)-3'), Poly(U)-binding-splicing factor PUF60, ... | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-10 | | Release date: | 2022-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of tandem domain RRM1-2 of FIR bound to FUSE ssDNA fragment
To Be Published
|
|
7QDE
 
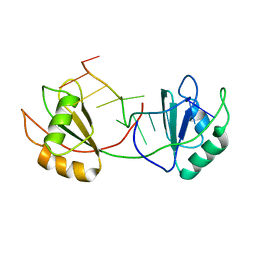 | |
7QR4
 
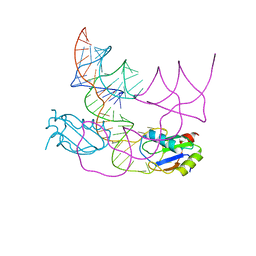 | | human CPEB3 HDV-like ribozyme | | Descriptor: | RNA CPEB3 ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Przytula-Mally, A.I, Engilberge, S, Johannsen, S, Olieric, V, Masquida, B, Sigel, R.K.O. | | Deposit date: | 2022-01-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Anticodon-like loop-mediated dimerization in the crystal structures of HdV-like CPEB3 ribozymes
Biorxiv, 2022
|
|
7QDD
 
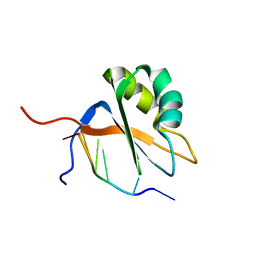 | |
7QR3
 
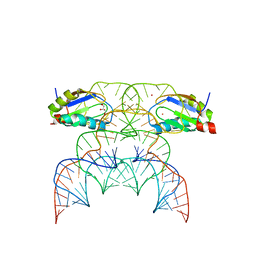 | | Chimpanzee CPEB3 HDV-like ribozyme | | Descriptor: | GLYCEROL, POTASSIUM ION, U1 small nuclear ribonucleoprotein A, ... | | Authors: | Przytula-Mally, A.I, Engilberge, S, Johannsen, S, Olieric, V, Masquida, B, Sigel, R.K.O. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Anticodon-like loop-mediated dimerization in the crystal structures of HdV-like CPEB3 ribozymes
Biorxiv, 2022
|
|
