1T10
 
 | |
1T11
 
 | | Trigger Factor | | Descriptor: | Trigger factor | | Authors: | Ludlam, A.V, Moore, B.A, Xu, Z. | | Deposit date: | 2004-04-14 | | Release date: | 2004-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of ribosomal chaperone trigger factor from Vibrio cholerae.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1T12
 
 | | Solution Structure of a new LTP1 | | Descriptor: | NONSPECIFIC LIPID-TRANSFER PROTEIN 1 | | Authors: | da Silva, P, Landon, C, Industri, B, Ponchet, M, Vovelle, F. | | Deposit date: | 2004-04-15 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a tobacco lipid transfer protein exhibiting
new biophysical and biological features
Proteins, 59, 2005
|
|
1T13
 
 | | Crystal Structure Of Lumazine Synthase From Brucella Abortus Bound To 5-nitro-6-(D-ribitylamino)-2,4(1H,3H) pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Klinke, S, Zylberman, V, Vega, D.R, Guimaraes, B.G, Braden, B.C, Goldbaum, F.A. | | Deposit date: | 2004-04-15 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic studies on Decameric Brucella spp. Lumazine Synthase: A Novel Quaternary Arrangement Evolved for a New Function?
J.Mol.Biol., 353, 2005
|
|
1T14
 
 | |
1T15
 
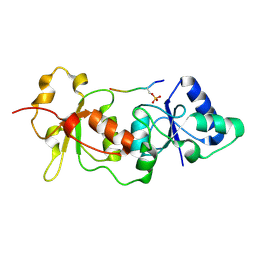 | | Crystal Structure of the Brca1 BRCT Domains in Complex with the Phosphorylated Interacting Region from Bach1 Helicase | | Descriptor: | BRCA1 interacting protein C-terminal helicase 1, Breast cancer type 1 susceptibility protein | | Authors: | Clapperton, J.A, Manke, I.A, Lowery, D.M, Ho, T, Haire, L.F, Yaffe, M.B, Smerdon, S.J. | | Deposit date: | 2004-04-15 | | Release date: | 2004-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T16
 
 | | Crystal structure of the bacterial fatty acid transporter FadL from Escherichia coli | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, COPPER (II) ION, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | van den Berg, B, Black, P.N, Clemons Jr, W.M, Rapoport, T.A. | | Deposit date: | 2004-04-15 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the long-chain fatty acid transporter FadL.
Science, 304, 2004
|
|
1T17
 
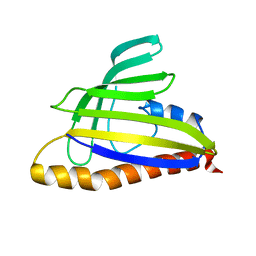 | | Solution Structure of the 18 kDa Protein CC1736 from Caulobacter crescentus: The Northeast Structural Genomics Consortium Target CcR19 | | Descriptor: | conserved hypothetical protein | | Authors: | Shen, Y, Atreya, H.S, Acton, T, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the 18 kDa protein CC1736 from Caulobacter crescentus identifies a member of the START domain superfamily and suggests residues mediating substrate specificity.
Proteins, 58, 2005
|
|
1T18
 
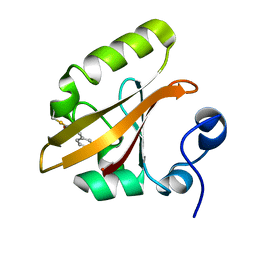 | | Early intermediate IE1 from time-resolved crystallography of the E46Q mutant of PYP | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Rajagopal, S, Anderson, S, Srajer, V, Schmidt, M, Pahl, R, Moffat, K. | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Structural Pathway for Signaling in the E46Q Mutant of Photoactive Yellow Protein
Structure, 13, 2005
|
|
1T19
 
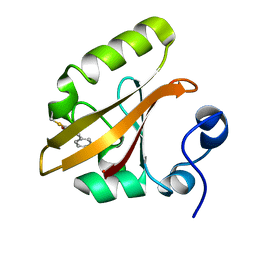 | | Early intermediate IE2 from time-resolved crystallography of the E46Q mutant of PYP | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Rajagopal, S, Anderson, S, Srajer, V, Schmidt, M, Pahl, R, Moffat, K. | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Structural Pathway for Signaling in the E46Q Mutant of Photoactive Yellow Protein
Structure, 13, 2005
|
|
1T1A
 
 | | Late intermediate IL1 from time-resolved crystallography of the E46Q mutant of PYP | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Rajagopal, S, Anderson, S, Srajer, V, Schmidt, M, Pahl, R, Moffat, K. | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Structural Pathway for Signaling in the E46Q Mutant of Photoactive Yellow Protein
Structure, 13, 2005
|
|
1T1B
 
 | | Late intermediate IL2 from time-resolved crystallography of the E46Q mutant of PYP | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Rajagopal, S, Anderson, S, Srajer, V, Schmidt, M, Pahl, R, Moffat, K. | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Structural Pathway for Signaling in the E46Q Mutant of Photoactive Yellow Protein
Structure, 13, 2005
|
|
1T1C
 
 | | Late intermediate IL3 from time-resolved crystallography of the E46Q mutant of PYP | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Rajagopal, S, Anderson, S, Srajer, V, Schmidt, M, Pahl, R, Moffat, K. | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Structural Pathway for Signaling in the E46Q Mutant of Photoactive Yellow Protein
Structure, 13, 2005
|
|
1T1D
 
 | |
1T1E
 
 | | High Resolution Crystal Structure of the Intact Pro-Kumamolisin, a Sedolisin Type Proteinase (previously called Kumamolysin or KSCP) | | Descriptor: | CALCIUM ION, kumamolisin | | Authors: | Comellas-Bigler, M, Maskos, K, Huber, R, Oyama, H, Oda, K, Bode, W. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | 1.2 a crystal structure of the serine carboxyl proteinase pro-kumamolisin: structure of an intact pro-subtilase
Structure, 12, 2004
|
|
1T1F
 
 | | Crystal Structure of Native Antithrombin in its Monomeric Form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III, GLYCEROL, ... | | Authors: | Johnson, D.J.D, Huntington, J.A. | | Deposit date: | 2004-04-16 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of monomeric native antithrombin reveals a novel reactive center loop conformation
J.Biol.Chem., 281, 2006
|
|
1T1G
 
 | | High Resolution Crystal Structure of Mutant E23A of Kumamolisin, a sedolisin type proteinase (previously called Kumamolysin or KSCP) | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin | | Authors: | Comellas-Bigler, M, Maskos, K, Huber, R, Oyama, H, Oda, K, Bode, W. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | 1.2 a crystal structure of the serine carboxyl proteinase pro-kumamolisin: structure of an intact pro-subtilase
Structure, 12, 2004
|
|
1T1H
 
 | | NMR solution structure of the U box domain from AtPUB14, an armadillo repeat containing protein from Arabidopsis thaliana | | Descriptor: | armadillo repeat containing protein | | Authors: | Andersen, P, Kragelund, B.B, Olsen, A.N, Larsen, F.H, Chua, N.-H, Poulsen, F.M, Skriver, K. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Biochemical Function of a Prototypical Arabidopsis U-box Domain
J.Biol.Chem., 279, 2004
|
|
1T1I
 
 | | High Resolution Crystal Structure of Mutant W129A of Kumamolisin, a Sedolisin Type Proteinase (previously called Kumamolysin or KSCP) | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin | | Authors: | Comellas-Bigler, M, Maskos, K, Huber, R, Oyama, H, Oda, K, Bode, W. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 1.2 a crystal structure of the serine carboxyl proteinase pro-kumamolisin: structure of an intact pro-subtilase
Structure, 12, 2004
|
|
1T1J
 
 | | Crystal structure of genomics APC5043 | | Descriptor: | hypothetical protein | | Authors: | Dong, A, Xu, X, Liu, Y, Zhang, R, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein PA1492 from Pseudomonas aeruginosa
TO BE PUBLISHED
|
|
1T1K
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT HIS-B10-ASP, VAL-B12-ALA, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin | | Authors: | Huang, K, Xu, B, Hu, S.Q, Chu, Y.C, Hua, Q.X, Whittaker, J, Nakagawa, S.H, De Meyts, P, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-10 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | How Insulin Binds: the B-Chain alpha-Helix Contacts the L1 beta-Helix of the Insulin Receptor.
J.Mol.Biol., 341, 2004
|
|
1T1L
 
 | | Crystal structure of the long-chain fatty acid transporter FadL | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Long-chain fatty acid transport protein | | Authors: | van den Berg, B, Black, P.N, Clemons Jr, W.M, Rapoport, T.A. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the long-chain fatty acid transporter FadL.
Science, 304, 2004
|
|
1T1M
 
 | | Binding position of ribosome recycling factor (RRF) on the E. coli 70S ribosome | | Descriptor: | 42-mer fragment of double helix from 16S rRNA, dodecamer fragment of double helix from 23S rRNA, ribosome recycling factor | | Authors: | Agrawal, R.K, Sharma, M.R, Kiel, M.C, Hirokawa, G, Booth, T.M, Spahn, C.M, Grassucci, R.A, Kaji, A, Frank, J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1T1N
 
 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN | | Descriptor: | CARBON MONOXIDE, PROTEIN (HEMOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mazzarella, L, Vitagliano, L, Savino, C, Zagari, A. | | Deposit date: | 1999-03-05 | | Release date: | 1999-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Trematomus newnesi haemoglobin re-opens the root effect question.
J.Mol.Biol., 287, 1999
|
|
1T1O
 
 | | Components of the control 70S ribosome to provide reference for the RRF binding site | | Descriptor: | 19-mer fragment of the 23S rRNA, 42-mer fragment of double helix from 16S rRNA, dodecamer fragment of double helix from 23S rRNA | | Authors: | Agrawal, R.K, Sharma, M.R, Kiel, M.C, Hirokawa, G, Booth, T.M, Spahn, C.M, Grassucci, R.A, Kaji, A, Frank, J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
