4K87
 
 | | Crystal structure of human prolyl-tRNA synthetase (substrate bound form) | | Descriptor: | ADENOSINE, PROLINE, Proline--tRNA ligase, ... | | Authors: | Hwang, K.Y, Son, J.H, Lee, E.H. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Conformational changes in human prolyl-tRNA synthetase upon binding of the substrates proline and ATP and the inhibitor halofuginone.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5HS1
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) complexed with Voriconazole | | Descriptor: | Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE, Voriconazole | | Authors: | Sabherwal, M, Sagatova, A, Keniya, M.V, Wilson, R.K, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2016-01-24 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14 alpha-demethylase.
Sci Rep, 6, 2016
|
|
7DBW
 
 | |
5HMM
 
 | | Crystal Structure of T5 D15 Protein Co-crystallized with Metal Ions | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exodeoxyribonuclease, ... | | Authors: | Flemming, C.S, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-16 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3M00
 
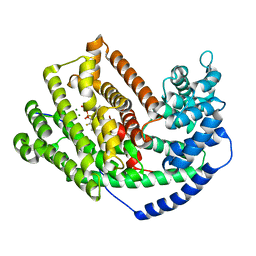 | | Crystal Structure of 5-epi-aristolochene synthase M4 mutant complexed with (2-cis,6-trans)-2-fluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, MAGNESIUM ION | | Authors: | Noel, J.P, Dellas, N, Faraldos, J.A, Zhao, M, Hess Jr, B.A, Smentek, L, Coates, R.M, O'Maille, P.E. | | Deposit date: | 2010-03-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural elucidation of cisoid and transoid cyclization pathways of a sesquiterpene synthase using 2-fluorofarnesyl diphosphates.
Acs Chem.Biol., 5, 2010
|
|
5HN5
 
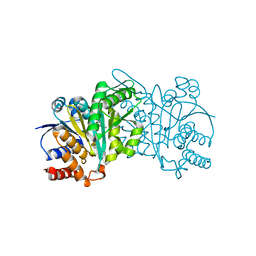 | | Crystal structure of beta-decarboxylating dehydrogenase (TK0280) from Thermococcus kodakarensis complexed with Mn and isocitrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Homoisocitrate dehydrogenase, ISOCITRIC ACID, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2016-01-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and function of an ancestral-type beta-decarboxylating dehydrogenase from Thermococcus kodakarensis
Biochem. J., 474, 2017
|
|
4XAZ
 
 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphotriesterase variant PTE-R18, ZINC ION | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
7DFO
 
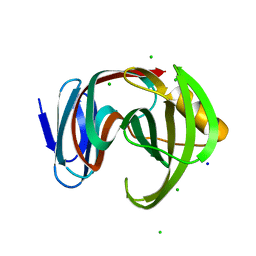 | | Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with 4-O-methyl-alpha-D-glucuronopyranosyl xylotetraose | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, CHLORIDE ION, ... | | Authors: | Fujimoto, Z, Kishine, N, Kaneko, S. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based substrate specificity analysis of GH11 xylanase from Streptomyces olivaceoviridis E-86.
Appl.Microbiol.Biotechnol., 105, 2021
|
|
1EXN
 
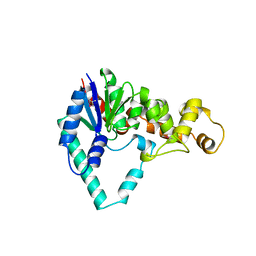 | | T5 5'-EXONUCLEASE | | Descriptor: | 5'-EXONUCLEASE | | Authors: | Ceska, T.A, Sayers, J.R, Stier, G, Suck, D. | | Deposit date: | 1997-01-17 | | Release date: | 1997-07-07 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A helical arch allowing single-stranded DNA to thread through T5 5'-exonuclease.
Nature, 382, 1996
|
|
3MPI
 
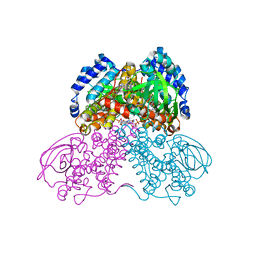 | | Structure of the glutaryl-coenzyme A dehydrogenase glutaryl-CoA complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, glutaryl-coenzyme A | | Authors: | Wischgoll, S, Warkentin, E, Boll, M, Ermler, U. | | Deposit date: | 2010-04-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for promoting and preventing decarboxylation in glutaryl-coenzyme a dehydrogenases.
Biochemistry, 49, 2010
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XD4
 
 | | Phosphotriesterase variant E2b | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R3, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4KDF
 
 | |
7DS7
 
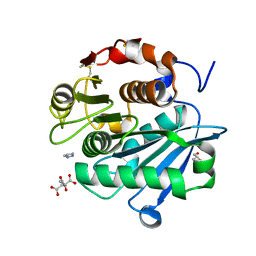 | | The Crystal Structure of Leaf-branch compost cutinase from Biortus. | | Descriptor: | CITRIC ACID, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wang, F, Lv, Z, Cheng, W, Lin, D, Chu, F, Xu, X, Tan, J. | | Deposit date: | 2020-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Leaf-branch compost cutinase from Biortus.
To Be Published
|
|
7DD3
 
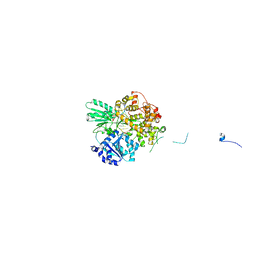 | | Cryo-EM structure of the pre-mRNA-loaded DEAH-box ATPase/helicase Prp2 in complex with Spp2 | | Descriptor: | PRP2 isoform 1, Pre-mRNA-splicing factor SPP2, pre-mRNA | | Authors: | Bai, R, Wan, R, Yan, C, Qi, J, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
3MSG
 
 | | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. | | Descriptor: | ACETATE ION, GLYCEROL, Intra-cellular xylanase ixt6, ... | | Authors: | Solomon, V, Zolotnitsky, G, Alhadeff, R, Shoham, Y, Shoham, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus.
TO BE PUBLISHED
|
|
5HWA
 
 | | Crystal Structure of MH-K1 chitosanase in substrate-bound form | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CACODYLATE ION, ... | | Authors: | Suzuki, M, Saito, A, Ando, A, Miki, K, Saito, J. | | Deposit date: | 2016-01-29 | | Release date: | 2017-02-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the GH-46 subclass III chitosanase from Bacillus circulans MH-K1 in complex with chitotetraose
Biomed.Biochim.Acta, 1868, 2024
|
|
4XES
 
 | | Structure of active-like neurotensin receptor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B.E, White, J.F, Shah, P, Grisshammer, R. | | Deposit date: | 2014-12-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural prerequisites for G-protein activation by the neurotensin receptor.
Nat Commun, 6, 2015
|
|
4KF2
 
 | | Structure of the P4509 BM3 A82F F87V heme domain | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leys, D. | | Deposit date: | 2013-04-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Key Mutations Alter the Cytochrome P450 BM3 Conformational Landscape and Remove Inherent Substrate Bias.
J.Biol.Chem., 288, 2013
|
|
1S9A
 
 | | Crystal Structure of 4-Chlorocatechol 1,2-dioxygenase from Rhodococcus opacus 1CP | | Descriptor: | (1-HEXADECANOYL-2-TETRADECANOYL-GLYCEROL-3-YL) PHOSPHONYL CHOLINE, BENZOIC ACID, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Solyanikova, I.P, Kolomytseva, M.P, Scozzafava, A, Golovleva, L.A, Briganti, F. | | Deposit date: | 2004-02-04 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of 4-chlorocatechol 1,2-dioxygenase from the chlorophenol-utilizing gram-positive Rhodococcus opacus 1CP.
J.Biol.Chem., 279, 2004
|
|
4KGV
 
 | | The R state structure of E. coli ATCase with ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, ... | | Authors: | Cockrell, G.M, Zheng, Y, Guo, W, Peterson, A.W, Kantrowitz, E.R. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New Paradigm for Allosteric Regulation of Escherichia coli Aspartate Transcarbamoylase.
Biochemistry, 52, 2013
|
|
3M02
 
 | | The Crystal Structure of 5-epi-aristolochene synthase complexed with (2-cis,6-trans)-2-fluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, MAGNESIUM ION | | Authors: | Noel, J.P, Dellas, N, Faraldos, J.A, Zhao, M, Hess Jr, B.A, Smentek, L, Coates, R.M, O'Maille, P.E. | | Deposit date: | 2010-03-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of cisoid and transoid cyclization pathways of a sesquiterpene synthase using 2-fluorofarnesyl diphosphates.
Acs Chem.Biol., 5, 2010
|
|
1EGD
 
 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
3M6I
 
 | | L-arabinitol 4-dehydrogenase | | Descriptor: | L-arabinitol 4-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2010-03-15 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and engineering of L-arabinitol 4-dehydrogenase from Neurospora crassa
J.Mol.Biol., 402, 2010
|
|
5HXZ
 
 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Barbiturase, CHLORIDE ION, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-01-31 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
