6HML
 
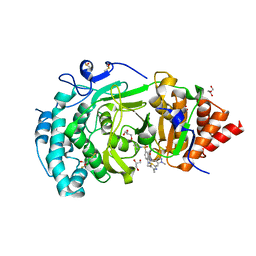 | | POLYADPRIBOSYL GLYCOSIDASE IN COMPLEX WITH PDD00017299 | | Descriptor: | 1-[(2,4-dimethyl-1,3-thiazol-5-yl)methyl]-6-[[(1-methylcyclopropyl)amino]-bis(oxidanyl)-$l^{4}-sulfanyl]-3-[(1-methylpyrazol-4-yl)methyl]quinazoline-2,4-dione, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tucker, J.A, Barkauskaite, E. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cell-Active Small Molecule Inhibitors of the DNA-Damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides.
J.Med.Chem., 61, 2018
|
|
6FXT
 
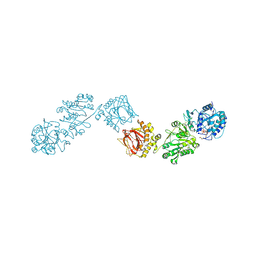 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Glc | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scietti, L, Chiapparino, A, De Giorgi, F, Fumagalli, M, Khoriauli, L, Nergadze, S, Basu, S, Olieric, V, Banushi, B, Giulotto, E, Gissen, P, Forneris, F. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun, 9, 2018
|
|
6BN8
 
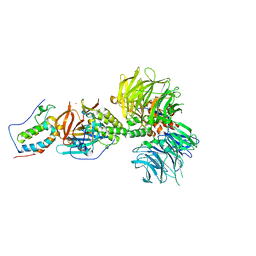 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET55 PROTAC. | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.990035 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
8TFP
 
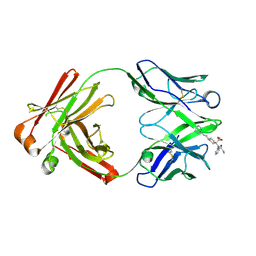 | | Fab from C10-S66K antibody in complex with carfentanil | | Descriptor: | Carfentanil, Heavy chain of Fab from C10-S66K antibody, Light chain of Fab from C10-S66K antibody | | Authors: | Pholcharee, T, Wilson, I.A. | | Deposit date: | 2023-07-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | An Engineered Human-Antibody Fragment with Fentanyl Pan-Specificity That Reverses Carfentanil-Induced Respiratory Depression.
Acs Chem Neurosci, 14, 2023
|
|
6Z4S
 
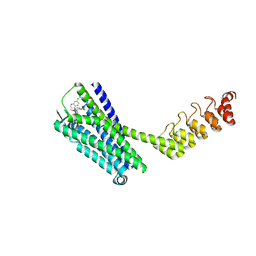 | | Crystal structure of the neurotensin receptor 1 (NTSR1-H4bmx) in complex with the small molecule inverse agonist SR48692 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,DARPin,HRV 3C protease recognition sequence | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
7KB0
 
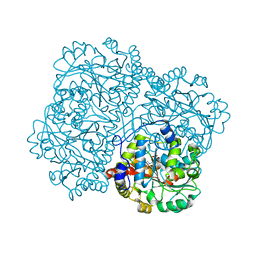 | | O-acety-L-homoserine aminocarboxypropyltransferase (MetY) from Thermotoga maritima with pyridoxal-5-phosphate (PLP) bound in the internal aldimine state | | Descriptor: | O-acetyl-L-homoserine sulfhydrylase | | Authors: | Brewster, J.L, Pachl, P, Squire, C, Selmer, M, Patrick, W.M. | | Deposit date: | 2020-10-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures and kinetics of Thermotoga maritima MetY reveal new insights into the predominant sulfurylation enzyme of bacterial methionine biosynthesis.
J.Biol.Chem., 296, 2021
|
|
6QZQ
 
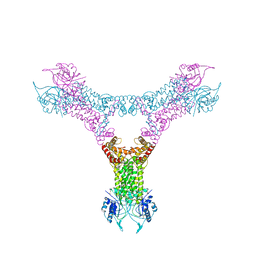 | |
6HFP
 
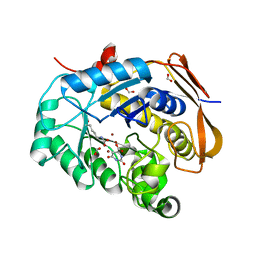 | |
6BIC
 
 | | 2.25 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | Descriptor: | (phenylmethyl) ~{N}-[(9~{S},12~{S},15~{S})-9-(hydroxymethyl)-12-(2-methylpropyl)-6,11,14-tris(oxidanylidene)-1,5,10,13,18,19-hexazabicyclo[15.2.1]icosa-17(20),18-dien-15-yl]carbamate, 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
8TMS
 
 | | Crystal structure of bacterial pectin methylesterase PmeC2 from rumen Butyrivibrio | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Pectinesterase | | Authors: | Carbone, V, Reilly, K, Sang, C, Schofield, L, Ronimus, R, Kelly, W.J, Attwood, G.T, Palevich, N. | | Deposit date: | 2023-07-30 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Bacterial Pectin Methylesterases Pme8A and PmeC2 from Rumen Butyrivibrio .
Int J Mol Sci, 24, 2023
|
|
6Z66
 
 | | Crystal structure of apo-state neurotensin receptor 1 | | Descriptor: | Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-27 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.192 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6G5N
 
 | | Crystal structure of human SP100 in complex with bromodomain-focused fragment XS039818e 1-(3-Phenyl-1,2,4-oxadiazol-5-yl)methanamine | | Descriptor: | (3-phenyl-1,2,4-oxadiazol-5-yl)methanamine, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Talon, R.P.H, Krojer, T, Tallant, C, Nunez-Alonso, G, Fairhead, M, Szykowska, A, Collins, P, Pearce, N.M, Ng, J, MacLean, E, Wright, N, Douangamath, A, Brandao-Neto, J, Burgess-Brown, N, Huber, K, Knapp, S, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2018-03-29 | | Release date: | 2018-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | Identifying small molecule binding sites for epigenetic proteins at domain-domain interfaces
Biorxiv, 2018
|
|
6BIJ
 
 | | HLA-DRB1 in complex with citrullinated fibrinogen peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Citrullinated Fibrinogen 72,74Cit69-81, HLA class II histocompatibility antigen, ... | | Authors: | Ting, Y.T, Scally, S.W, Rossjohn, J. | | Deposit date: | 2017-11-02 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The interplay between citrullination and HLA-DRB1 polymorphism in shaping peptide binding hierarchies in rheumatoid arthritis.
J. Biol. Chem., 293, 2018
|
|
7KEF
 
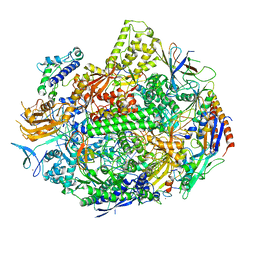 | | RNA polymerase II elongation complex with unnatural base dTPT3, rNaM in swing state | | Descriptor: | (1S)-1,4-anhydro-1-(3-methoxynaphthalen-2-yl)-5-O-phosphono-D-ribitol, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
6ZIL
 
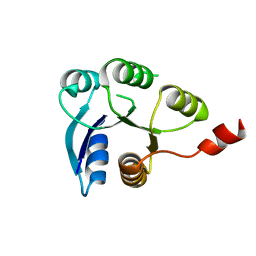 | |
6R0E
 
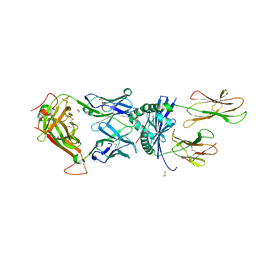 | |
6G6E
 
 | |
6ZA8
 
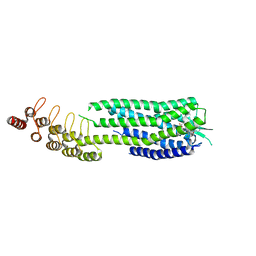 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule partial agonist RTI-3a | | Descriptor: | (2~{S})-2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]-4-methyl-pentanoic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1),Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1) | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
7K76
 
 | |
6HMY
 
 | |
8TTK
 
 | | Tryptophan-6-halogenase BorH apo structure | | Descriptor: | SULFATE ION, Tryptophan 6-halogenase | | Authors: | Lingkon, K, Bellizzi, J.J. | | Deposit date: | 2023-08-14 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic and thermodynamic evidence of negative cooperativity of flavin and tryptophan binding in the flavin-dependent halogenases AbeH and BorH.
Biorxiv, 2023
|
|
6Z4V
 
 | | Crystal structure of the neurotensin receptor 1 (NTSR1-H4bmx) in complex with NTS8-13 | | Descriptor: | ARG-PRO-TYR-ILE-LEU, Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6BIQ
 
 | |
5HCZ
 
 | | EGFR kinase domain mutant "TMLR" with 3-azetidinyl azaindazole compound 21 | | Descriptor: | 2-[1-[1-[(2~{S})-butan-2-yl]-6-[[2-(1-cyclopropylsulfonylpyrazol-4-yl)pyrimidin-4-yl]amino]pyrazolo[4,3-c]pyridin-3-yl]azetidin-3-yl]propan-2-ol, Epidermal growth factor receptor, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2016-01-04 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Discovery of a Noncovalent, Mutant-Selective Epidermal Growth Factor Receptor Inhibitor.
J.Med.Chem., 59, 2016
|
|
6HNS
 
 | |
