1S1X
 
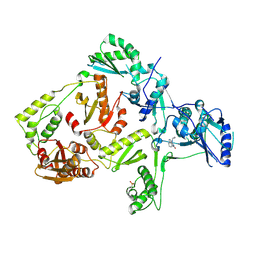 | | Crystal structure of V108I mutant HIV-1 reverse transcriptase in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
4WE1
 
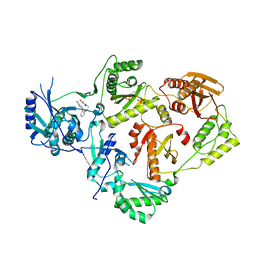 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ600) | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}naphthalene-2-carbonitrile, Gag-Pol polyprotein, MAGNESIUM ION, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Picomolar Inhibitors of HIV-1 Reverse Transcriptase: Design and Crystallography of Naphthyl Phenyl Ethers.
Acs Med.Chem.Lett., 5, 2014
|
|
6WCQ
 
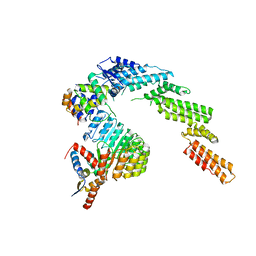 | | Structure of a substrate-bound DQC ubiquitin ligase | | Descriptor: | Cullin-1, F-box/LRR-repeat protein 17, Kelch-like ECH-associated protein 1, ... | | Authors: | Mena, E.L, Jevtic, P, Greber, B.J, Gee, C.L, Lew, B.G, Akopian, D, Nogales, E, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-31 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
1S1V
 
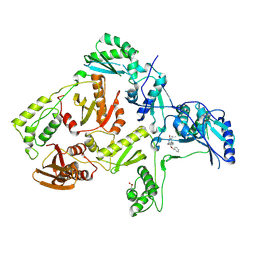 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with TNK-651 | | Descriptor: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
5EAP
 
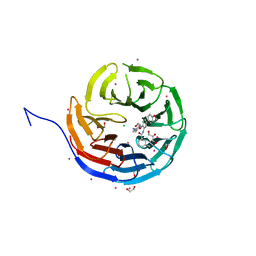 | | Crystal structure of human WDR5 in complex with compound 9e | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
6WXB
 
 | |
5EAR
 
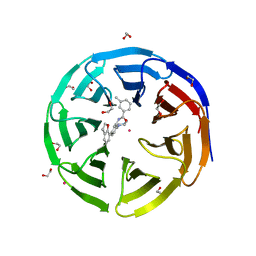 | | Crystal structure of human WDR5 in complex with compound 9d | | Descriptor: | 1,2-ETHANEDIOL, N-[5-(2,3-dihydro-1-benzofuran-7-yl)-2-(4-methylpiperazin-1-yl)phenyl]-3-methylbenzamide, SULFATE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
5ECJ
 
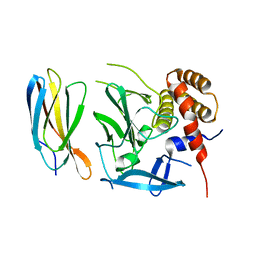 | |
1S1W
 
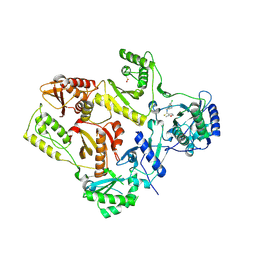 | | Crystal structure of V106A mutant HIV-1 reverse transcriptase in complex with UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1CL3
 
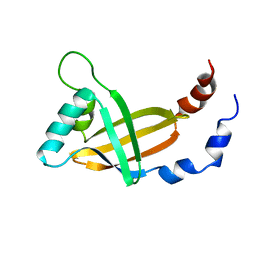 | | MOLECULAR INSIGHTS INTO PEBP2/CBF-SMMHC ASSOCIATED ACUTE LEUKEMIA REVEALED FROM THE THREE-DIMENSIONAL STRUCTURE OF PEBP2/CBF BETA | | Descriptor: | POLYOMAVIRUS ENHANCER BINDING PROTEIN 2 | | Authors: | Goger, M, Gupta, V, Kim, W.Y, Shigesada, K, Ito, Y, Werner, M.H. | | Deposit date: | 1999-05-04 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Molecular insights into PEBP2/CBF beta-SMMHC associated acute leukemia revealed from the structure of PEBP2/CBF beta
Nat.Struct.Biol., 6, 1999
|
|
1S1U
 
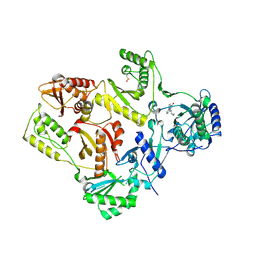 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
4V2S
 
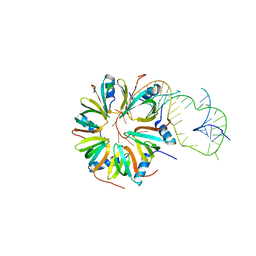 | | Crystal structure of Hfq in complex with the sRNA RydC | | Descriptor: | RNA-BINDING PROTEIN HFQ, RYDC | | Authors: | Dimastrogiovanni, D, Frohlich, K.S, Bruce, H.A, Bandyra, K.J, Hohensee, S, Vogel, J, Luisi, B.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Recognition of the small regulatory RNA RydC by the bacterial Hfq protein.
Elife, 3, 2014
|
|
5DOX
 
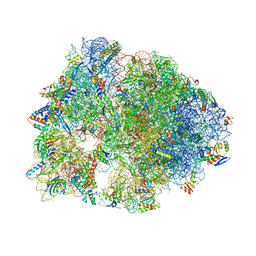 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Hygromycin-A at 3.1A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
2X0W
 
 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO 5,6-dimethoxy- 2-methylbenzothiazole | | Descriptor: | 5,6-DIMETHOXY-2-METHYL-1,3-BENZOTHIAZOLE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Kaar, J.L, Basse, N, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
5NIM
 
 | | EthR complex | | Descriptor: | HTH-type transcriptional regulator EthR, SULFATE ION, [1-(2-hydroxyethyl)pyrrolo[3,4-c]pyrazol-5-yl]-(5-propyl-1,2-oxazol-3-yl)methanone | | Authors: | Pohl, E, Tatum, N.J, Cole, J.C, Baulard, A.R. | | Deposit date: | 2017-03-24 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New active leads for tuberculosis booster drugs by structure-based drug discovery.
Org. Biomol. Chem., 15, 2017
|
|
5NJ0
 
 | | EthR complex | | Descriptor: | HTH-type transcriptional regulator EthR, SULFATE ION, ~{N}-[(2~{R})-2-(4-nitrophenyl)-2-oxidanyl-ethyl]-1,3-benzodioxole-5-carboxamide | | Authors: | Pohl, E, Tatum, N.J, Cole, J.C, Baulard, A.R. | | Deposit date: | 2017-03-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New active leads for tuberculosis booster drugs by structure-based drug discovery.
Org. Biomol. Chem., 15, 2017
|
|
2X0U
 
 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO A 2-amino substituted benzothiazole scaffold | | Descriptor: | 6,7-DIHYDRO[1,4]DIOXINO[2,3-F][1,3]BENZOTHIAZOL-2-AMINE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Kaar, J.L, Basse, N, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
2X0V
 
 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO 4-(trifluoromethyl)benzene-1,2-diamine | | Descriptor: | 4-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Basse, N, Kaar, J.L, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
3LAL
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with N1-ethyl pyrimidinedione non-nucleoside inhibitor | | Descriptor: | 3-{[3-ethyl-5-(1-methylethyl)-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl]carbonyl}-5-methylbenzonitrile, HIV Reverse transcriptase, SULFATE ION | | Authors: | Lansdon, E.B, Mitchell, M.L. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | N1-Alkyl pyrimidinediones as non-nucleoside inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3CKC
 
 | | B. thetaiotaomicron SusD | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
3CVI
 
 | |
3L7U
 
 | | Crystal structure of human NM23-H1 | | Descriptor: | Nucleoside diphosphate kinase A, PHOSPHATE ION | | Authors: | Han, B.G, Min, K, Lee, B.I, Lee, S. | | Deposit date: | 2009-12-29 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined Structure of Human NM23-H1 from a Hexagonal Crystal
BULL.KOREAN CHEM.SOC., 31, 2010
|
|
3LAK
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with N1-heterocycle pyrimidinedione non-nucleoside inhibitor | | Descriptor: | 3-({3-[(2-amino-6-fluoropyridin-4-yl)methyl]-5-(1-methylethyl)-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl}carbonyl)-5-methylbenzonitrile, CHLORIDE ION, HIV Reverse transcriptase, ... | | Authors: | Lansdon, E.B, Mitchell, M.L. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N1-Heterocyclic pyrimidinediones as non-nucleoside inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3CMA
 
 | |
4FHI
 
 | | Development of synthetically accessible non-secosteroidal hybrid molecules combining vitamin D receptor agonism and histone deacetylase inhibition | | Descriptor: | N-hydroxy-2-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}acetamide, SRC-1, Vitamin Nuclear Receptor | | Authors: | Fischer, J, Wang, T.T, Kaldre, D, Rochel, N, Moras, D, White, J.H, Gleason, J.L. | | Deposit date: | 2012-06-06 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthetically accessible non-secosteroidal hybrid molecules combining vitamin d receptor agonism and histone deacetylase inhibition.
Chem.Biol., 19, 2012
|
|
