3IYD
 
 | | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DNA (98-MER), ... | | Authors: | Hudson, B.P, Quispe, J, Lara, S, Kim, Y, Berman, H, Arnold, E, Ebright, R.H, Lawson, C.L. | | Deposit date: | 2009-08-01 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (19.799999 Å) | | Cite: | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5DDK
 
 | | Succinyl-CoA:acetate CoA-transferase (AarCH6-N347A) in complex with CoA | | Descriptor: | Acetyl-CoA hydrolase, CHLORIDE ION, COENZYME A, ... | | Authors: | Kappock, T.J, Mullins, E.A, Murphy, J.R. | | Deposit date: | 2015-08-25 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Functional Dissection of the Bipartite Active Site of the Class I Coenzyme A (CoA)-Transferase Succinyl-CoA:Acetate CoA-Transferase.
Front Chem, 4, 2016
|
|
5XQ5
 
 | |
1PZJ
 
 | | Cholera Toxin B-Pentamer Complexed With Nitrophenyl Galactoside 5 | | Descriptor: | Cholera Toxin B Subunit, N-{3-[4-(3-AMINO-PROPYL)-PIPERAZIN-1-YL]-PROPYL}-3-NITRO-5-(GALACTOPYRANOSYL)-ALPHA-BENZAMIDE, N-{3-[4-(3-AMINO-PROPYL)-PIPERAZIN-1-YL]-PROPYL}-3-NITRO-5-(GALACTOPYRANOSYL)-BETA-BENZAMIDE | | Authors: | Mitchell, D.D, Pickens, J.C, Korotkov, K, Fan, E, Hol, W.G.J. | | Deposit date: | 2003-07-11 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | 3,5-Substituted phenyl galactosides as leads in designing effective cholera toxin antagonists; synthesis and crystallographic studies
Bioorg.Med.Chem., 12, 2004
|
|
5NKW
 
 | | X-ray crystal structure of an AA9 LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Frandsen, K.E.H, Poulsen, J.-C.N, Lo Leggio, L. | | Deposit date: | 2017-04-03 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and electronic determinants of lytic polysaccharide monooxygenase reactivity on polysaccharide substrates.
Nat Commun, 8, 2017
|
|
2HCJ
 
 | | Trypsin-modified Elongation Factor Tu in complex with tetracycline | | Descriptor: | GLYOXYLIC ACID, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mui, S, Heffron, S.E, Aorora, A, Abel, K, Bergmann, E, Jurnak, F. | | Deposit date: | 2006-06-16 | | Release date: | 2006-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Molecular complementarity between tetracycline and the GTPase active site of elongation factor Tu.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2OBY
 
 | | Crystal structure of Human P53 inducible oxidoreductase (TP53I3,PIG3) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative quinone oxidoreductase | | Authors: | Porte, S, Valencia, E, Farres, J, Fita, I, Pike, A.C.W, Shafqat, N, Debreczeni, J, Johansson, C, Haroniti, A, Gileadi, O, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, von Delft, F, Oppermann, U, Pares, X. | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human P53 inducible oxidoreductase (TP53I3, PIG3)
To be Published
|
|
3E1L
 
 | | Crystal structure of E. coli Bacterioferritin (BFR) soaked in phosphate with an alternative conformation of the unoccupied Ferroxidase centre (APO-BFR II). | | Descriptor: | BACTERIOFERRITIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Crow, A, Lawson, T, Lewin, A, Moore, G.R, Le Brun, N. | | Deposit date: | 2008-08-04 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for iron mineralization by bacterioferritin
J.Am.Chem.Soc., 131, 2009
|
|
6BFM
 
 | |
6BG0
 
 | | Caspase-3 Mutant - D9A,D28A,S150D | | Descriptor: | AC-ASP-GLU-VAL-ASP-CMK, AZIDE ION, Caspase-3, ... | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-27 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.125 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
5NBD
 
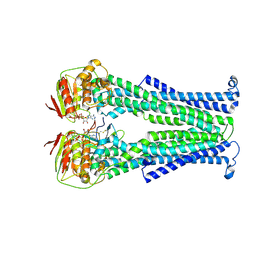 | | PglK flippase in complex with inhibitory nanobody | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nanobody, WlaB protein | | Authors: | Perez, C, Pardon, E, Steyaert, J, Locher, K.P. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural basis of inhibition of lipid-linked oligosaccharide flippase PglK by a conformational nanobody.
Sci Rep, 7, 2017
|
|
5CZ8
 
 | |
5D0Z
 
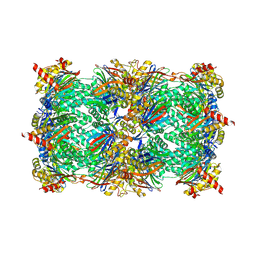 | |
5NO1
 
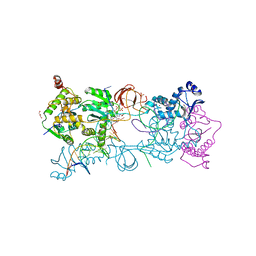 | |
2OGA
 
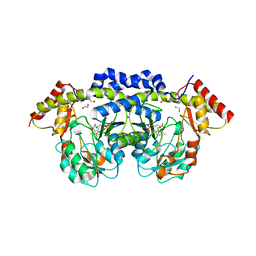 | |
5SX5
 
 | |
5N02
 
 | | Crystal structure of the decarboxylase AibA/AibB C56S variant | | Descriptor: | ACETATE ION, Glutaconate CoA-transferase family, subunit A, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5CZ7
 
 | |
2GRZ
 
 | | 5ns Photoproduct of the M37V mutant of Scapharca HbI | | Descriptor: | CARBON MONOXIDE, Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Pahl, R, Srajer, V, Royer Jr, W.E. | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Allosteric action in real time: Time-resolved crystallographic studies of a cooperative dimeric hemoglobin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5D0X
 
 | | Yeast 20S proteasome beta5-T1S mutant in complex with Bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-08-03 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
1ODI
 
 | |
5N5W
 
 | | 14-3-3 sigma in complex with TAZ pS89 peptide and fragment NV3 | | Descriptor: | 14-3-3 protein sigma, 4-[3,5-bis(chloranyl)pyridin-2-yl]oxyaniline, CALCIUM ION, ... | | Authors: | Sijbesma, E, Leysen, S, Ottmann, C. | | Deposit date: | 2017-02-14 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Identification of Two Secondary Ligand Binding Sites in 14-3-3 Proteins Using Fragment Screening.
Biochemistry, 56, 2017
|
|
4EAH
 
 | | Crystal structure of the formin homology 2 domain of FMNL3 bound to actin | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Thompson, M.E, Heimsath, E.G, Gauvin, T.J, Higgs, H.N, Kull, F.J. | | Deposit date: | 2012-03-22 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | FMNL3 FH2-actin structure gives insight into formin-mediated actin nucleation and elongation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4IHA
 
 | | Crystal structure of rice DWARF14 (D14) in complex with a GR24 hydrolysis intermediate | | Descriptor: | (2R,3R)-2,4,4-trihydroxy-3-methylbutanal, Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
6CH0
 
 | | Structure of the Quorum Quenching lactonase from Alicyclobacillus acidoterrestris bound to a glycerol molecule | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, COBALT (II) ION, ... | | Authors: | Bergonzi, C, Schwab, M, Naik, T, Daude, D, Chabriere, E, Elias, M. | | Deposit date: | 2018-02-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Biochemical Characterization of AaL, a Quorum Quenching Lactonase with Unusual Kinetic Properties.
Sci Rep, 8, 2018
|
|
