7M8E
 
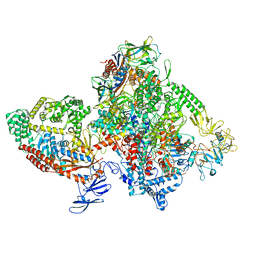 | | E.coli RNAP-RapA elongation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, W, Liu, B. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for activation of Swi2/Snf2 ATPase RapA by RNA polymerase.
Nucleic Acids Res., 49, 2021
|
|
8JNO
 
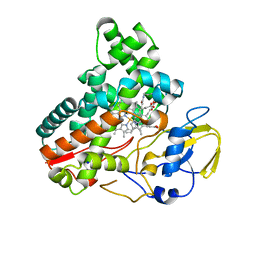 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate 10-epi-deOH-HSAF | | Descriptor: | (1Z,3E,5S,7S,8R,9S,10S,11R,13R,15R,16S,18Z,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.09,13.08,15.05,16]octacosa-1(2),3,18-triene-20,27,28-trione, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6V0Z
 
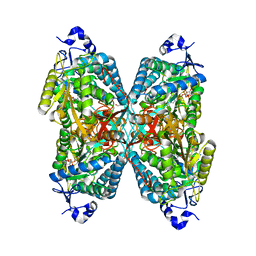 | | Structure of ALDH7A1 mutant R441C complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Korasick, D.A, Tanner, J.J. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biochemical, structural, and computational analyses of two new clinically identified missense mutations of ALDH7A1.
Chem.Biol.Interact., 2024
|
|
8EH2
 
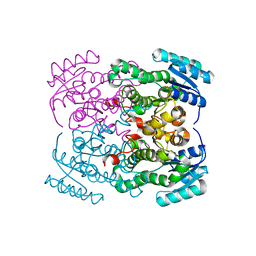 | |
6QX0
 
 | | HEWL lysozyme, crystallized from LiCl solution | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
6QX6
 
 | | Structure of gtPebB-dihydrobiliverdin complex | | Descriptor: | 15,16-DIHYDROBILIVERDIN, Ferredoxin bilin reductase plastid, PENTAETHYLENE GLYCOL, ... | | Authors: | Sommerkamp, J.A, Hofmann, E. | | Deposit date: | 2019-03-07 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the first eukaryotic bilin reductaseGtPEBB reveals a flipped binding mode of dihydrobiliverdin.
J.Biol.Chem., 294, 2019
|
|
3MHY
 
 | | A New PII Protein Structure | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-OXOGLUTARIC ACID, ... | | Authors: | Winkler, F.K, Truan, D, Li, X.D. | | Deposit date: | 2010-04-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A new P(II) protein structure identifies the 2-oxoglutarate binding site.
J.Mol.Biol., 400, 2010
|
|
6V6Y
 
 | | Crystal Structure of T. thermophilus methylenetetrahydrofolate dehydrogenase (MTHFD) | | Descriptor: | Bifunctional protein FolD, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Maiello, F, Coelho, C, Gallo, G, Sucharski, F, Hardy, L, Wurtele, M. | | Deposit date: | 2019-12-06 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15201569 Å) | | Cite: | Crystal structure of Thermus thermophilus methylenetetrahydrofolate dehydrogenase and determinants of thermostability.
Plos One, 15, 2020
|
|
1R5N
 
 | | Crystal Structure Analysis of sup35 complexed with GDP | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
8EW4
 
 | | Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 1 | | Descriptor: | COBALT (II) ION, MYRISTIC ACID, Serum albumin | | Authors: | Gucwa, M, Cooper, D.R, Unciano, J, Lea, K, Kim, L, Lenkiewicz, J, Starban, I, Stewart, A.J, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterisation of Co2+-binding sites on serum albumins and their interplay with fatty acids
Chem Sci, 14, 2023
|
|
8EY5
 
 | | Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 3 | | Descriptor: | COBALT (II) ION, MYRISTIC ACID, Serum albumin | | Authors: | Gucwa, M, Cooper, D.R, Stewart, A.J, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and biochemical characterisation of Co2+-binding sites on serum albumins and their interplay with fatty acids
Chem Sci, 14, 2023
|
|
6V7N
 
 | |
7M7D
 
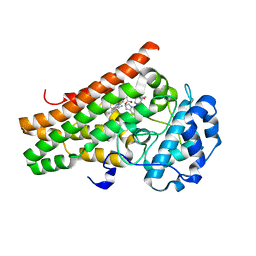 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with IACS-8968 | | Descriptor: | (5S)-6,6-dimethyl-8-[(4S)-7-(trifluoromethyl)imidazo[1,5-a]pyridin-5-yl]-1,3,8-triazaspiro[4.5]decane-2,4-dione, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leonard, P.G, Cross, J.B. | | Deposit date: | 2021-03-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
8C6I
 
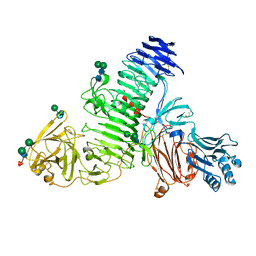 | | TMEM2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hohenester, E. | | Deposit date: | 2023-01-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the transmembrane protein 2 (TMEM2) ectodomain and its apparent lack of hyaluronidase activity.
Wellcome Open Res, 8, 2023
|
|
1RAM
 
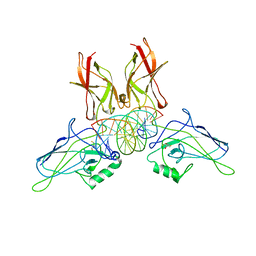 | | A NOVEL DNA RECOGNITION MODE BY NF-KB P65 HOMODIMER | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA (5'-D(*CP*GP*GP*CP*TP*GP*GP*AP*AP*AP*TP*TP*TP*CP*CP*AP*GP*CP*CP*G)-3'), PROTEIN (TRANSCRIPTION FACTOR NF-KB P65) | | Authors: | Chen, Y.-Q, Ghosh, S, Ghosh, G. | | Deposit date: | 1997-11-22 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel DNA recognition mode by the NF-kappa B p65 homodimer.
Nat.Struct.Biol., 5, 1998
|
|
7M63
 
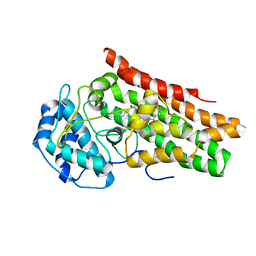 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with IACS-70099 | | Descriptor: | (2R)-N-(4-chlorophenyl)-2-[(1R,3S,5S,6r)-3-(5,6-difluoro-1H-benzimidazol-1-yl)bicyclo[3.1.0]hexan-6-yl]propanamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Leonard, P.G, Cross, J.B. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
2VUN
 
 | | The Crystal Structure of Enamidase at 1.9 A Resolution - A new Member of the Amidohydrolase Superfamily | | Descriptor: | CHLORIDE ION, ENAMIDASE, FE (III) ION, ... | | Authors: | Kress, D, Alhapel, A, Pierik, A.J, Essen, L.-O. | | Deposit date: | 2008-05-27 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Crystal Structure of Enamidase: A Bifunctional Enzyme of the Nicotinate Catabolism.
J.Mol.Biol., 384, 2008
|
|
5UXF
 
 | | Crystal Structure of mouse RECON (AKR1C13) in complex with Cyclic di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Dihydrodiol dehydrogenase | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2017-02-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Sensing of Bacterial Cyclic Dinucleotides by the Oxidoreductase RECON Promotes NF-kappa B Activation and Shapes a Proinflammatory Antibacterial State.
Immunity, 46, 2017
|
|
5FOU
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH FHPA TETRAPEPTIDE | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, FHPA PEPTIDE, PHOSPHATE ION | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
8EVU
 
 | | Cryo EM structure of Vibrio cholerae NQR | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit A, ... | | Authors: | Fuller, J.R, Juarez, O. | | Deposit date: | 2022-10-20 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.5804 Å) | | Cite: | Novel cofactor binding motifs, electron transfer and ion pumping mechanisms of the respiratory complex NQR
To Be Published
|
|
6SBP
 
 | |
6SBV
 
 | | X-ray Structure of Human LDH-A with an Allosteric Inhibitor (Compound 7) | | Descriptor: | L-lactate dehydrogenase A chain, ~{N}-[3-[(7-nitrodibenzofuran-2-yl)sulfonylamino]phenyl]-1-oxidanyl-cyclopropane-1-carboxamide | | Authors: | Friberg, A, Puetter, V, Nguyen, D, Rehwinkel, H. | | Deposit date: | 2019-07-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Evidence for Isoform-Selective Allosteric Inhibition of Lactate Dehydrogenase A.
Acs Omega, 5, 2020
|
|
8EW7
 
 | | Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 2 | | Descriptor: | COBALT (II) ION, MYRISTIC ACID, Serum albumin | | Authors: | Gucwa, M, Cooper, D.R, Stewart, A.J, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and biochemical characterisation of Co2+-binding sites on serum albumins and their interplay with fatty acids
Chem Sci, 14, 2023
|
|
6V4S
 
 | |
6VBY
 
 | | Cinnamate 4-hydroxylase (C4H1) from Sorghum bicolor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cinnamic acid 4-hydroxylase, GLYCEROL, ... | | Authors: | Zhang, B, Kang, C, Lewis, K.M. | | Deposit date: | 2019-12-19 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Function of the Cytochrome P450 Monooxygenase Cinnamate 4-hydroxylase fromSorghum bicolor.
Plant Physiol., 183, 2020
|
|
